From a52fee4bd8166fa22d7e7d042b73280e3eb1db65 Mon Sep 17 00:00:00 2001
From: arq5x <arq5x@virginia.edu>
Date: Wed, 19 Nov 2014 17:32:23 -0500
Subject: [PATCH] added tutorial
---
tutorial/Makefile | 3 +
tutorial/answers.html | 144 ++
tutorial/answers.md | 136 ++
tutorial/bedtools.html | 542 ++++
tutorial/bedtools.md | 715 ++++++
tutorial/bootstrap.css | 4480 ++++++++++++++++++++++++++++++++++
tutorial/template.class.html | 65 +
7 files changed, 6085 insertions(+)
create mode 100644 tutorial/Makefile
create mode 100644 tutorial/answers.html
create mode 100644 tutorial/answers.md
create mode 100644 tutorial/bedtools.html
create mode 100644 tutorial/bedtools.md
create mode 100644 tutorial/bootstrap.css
create mode 100644 tutorial/template.class.html
diff --git a/tutorial/Makefile b/tutorial/Makefile
new file mode 100644
index 00000000..d707406b
--- /dev/null
+++ b/tutorial/Makefile
@@ -0,0 +1,3 @@
+all:
+ pandoc --css bootstrap.css --template template.class.html -s -S bedtools.md -o bedtools.html
+ pandoc --css bootstrap.css --template template.class.html -s -S answers.md -o answers.html
diff --git a/tutorial/answers.html b/tutorial/answers.html
new file mode 100644
index 00000000..769de7c7
--- /dev/null
+++ b/tutorial/answers.html
@@ -0,0 +1,144 @@
+<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
+<html xmlns="http://www.w3.org/1999/xhtml">
+<head>
+ <meta http-equiv="Content-Type" content="text/html; charset=utf-8" />
+ <meta http-equiv="Content-Style-Type" content="text/css" />
+ <meta name="generator" content="pandoc" />
+ <meta name="author" content="Aaron Quinlan" />
+ <title>bedtools Tutorial</title>
+ <style type="text/css">code{white-space: pre;}</style>
+ <link rel="stylesheet" href="bootstrap.css" type="text/css" />
+</head>
+<body>
+ <div class="navbar navbar-static-top">
+ <div class="navbar-inner">
+ <div class="container">
+ <span class="doc-title">bedtools Tutorial</span>
+ <ul class="nav pull-right doc-info">
+ <li><p class="navbar-text">Aaron Quinlan</p></li>
+ </ul>
+ </div>
+ </div>
+ </div>
+ <div class="container">
+ <div class="row">
+ <div class="span12">
+ <h1 id="puzzles-to-help-teach-you-more-bedtools.">Puzzles to help teach you more bedtools.</h1>
+<ol style="list-style-type: decimal">
+<li>Create a BED file representing all of the intervals in the genome that are NOT exonic.</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools complement -i exons.bed -g genome.txt > notexons.bed</code></pre>
+<ol start="2" style="list-style-type: decimal">
+<li>What is the average distance from GWAS SNPs to the closest exon? (Hint - have a look at the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/closest.html">closest</a> tool.)</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools closest -a gwas.bed -b exons.bed -d | head
+chr1 1005805 1005806 rs3934834 chr1 1007125 1007955 NM_001205252_exon_0_0_chr1_1007126_r 0 - 1320
+chr1 1079197 1079198 rs11260603 chr1 1078118 1079434 NR_038869_exon_2_0_chr1_1078119_f 0 + 0
+chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256456_exon_1_0_chr1_1247398_r 0 - 0
+chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256460_exon_1_0_chr1_1247398_r 0 - 0
+chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256462_exon_1_0_chr1_1247398_r 0 - 0
+chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256463_exon_1_0_chr1_1247398_r 0 - 0
+chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_017871_exon_1_0_chr1_1247398_r 0 - 0
+chr1 2069171 2069172 rs425277 chr1 2066700 2066786 NM_001033581_exon_1_0_chr1_2066701_f 0 + 2386
+chr1 2069171 2069172 rs425277 chr1 2066700 2066786 NM_001033582_exon_1_0_chr1_2066701_f 0 + 2386
+chr1 2069171 2069172 rs425277 chr1 2066700 2066786 NM_001242874_exon_1_0_chr1_2066701_f 0 + 2386
+
+bedtools closest -a gwas.bed -b exons.bed -d \
+ | awk '{ sum += $11 } END { if (NR > 0) print sum / NR }'
+46713.1</code></pre>
+<ol start="3" style="list-style-type: decimal">
+<li>Count how many exons occur in each 500kb interval (“windowâ€) in the human genome. (Hint - have a look at the <code>makewindows</code> tool.)</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools makewindows -g genome.txt -w 500000 > genome.windows.bed
+bedtools intersect -a genome.windows.bed -b exons.bed -c > genome.windows.exoncount.bedg</code></pre>
+<p>or…</p>
+<pre><code>bedtools makewindows -g genome.txt -w 500000 \
+ | bedtools intersect -a - -b exons.bed -c \
+> genome.windows.exoncount.bedg</code></pre>
+<ol start="4" style="list-style-type: decimal">
+<li>Are there any exons that are completely overlapped by an enhancer? If so, how many?</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools intersect -a exons.bed \
+ -b <(grep Enhancer hesc.chromHmm.bed) \
+ -wa -wb -f 1.0 \
+| head
+chr1 948846 948956 NM_005101_exon_0_0_chr1_948847_f 0 + chr1 948337 949337 4_Strong_Enhancer
+chr1 1051439 1051736 NM_017891_exon_9_0_chr1_1051440_r 0 - chr1 1051337 1051737 6_Weak_Enhancer
+chr1 1109285 1109306 NM_001130045_exon_0_0_chr1_1109286_f 0 + chr1 1108537 1109537 6_Weak_Enhancer
+chr1 1109803 1109869 NM_001130045_exon_2_0_chr1_1109804_f 0 + chr1 1109737 1109937 6_Weak_Enhancer
+chr1 1219357 1219470 NM_001130413_exon_4_0_chr1_1219358_f 0 + chr1 1219137 1220137 7_Weak_Enhancer
+chr1 1219357 1219470 NR_037668_exon_4_0_chr1_1219358_f 0 + chr1 1219137 1220137 7_Weak_Enhancer
+chr1 1229202 1229313 NM_030649_exon_1_0_chr1_1229203_r 0 - chr1 1228937 1229937 6_Weak_Enhancer
+chr1 1229469 1229579 NM_030649_exon_2_0_chr1_1229470_r 0 - chr1 1228937 1229937 6_Weak_Enhancer
+chr1 1234724 1234736 NM_030649_exon_14_0_chr1_1234725_r 0 - chr1 1234137 1234937 7_Weak_Enhancer
+chr1 1245060 1245231 NM_153339_exon_4_0_chr1_1245061_f 0 + chr1 1244937 1245337 4_Strong_Enhancer
+
+bedtools intersect -a exons.bed \
+ -b <(grep Enhancer hesc.chromHmm.bed) \
+ -wa -wb -f 1.0 -u \
+| wc -l
+13746</code></pre>
+<ol start="5" style="list-style-type: decimal">
+<li>What fraction of the GWAS SNPs are exonic?</li>
+</ol>
+<p>Answer:</p>
+<pre><code>wc -l gwas.bed
+17680 gwas.bed
+
+bedtools intersect -a gwas.bed -b exons.bed -u | wc -l
+1625
+
+echo "foo" | awk '{print 1625/17680}'
+0.0919118</code></pre>
+<ol start="6" style="list-style-type: decimal">
+<li>What fraction of the GWAS SNPs are lie in either enhancers or promoters in the hESC data we have?</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools intersect -a gwas.bed -b <(egrep "Enhancer|Promoter" hesc.chromHmm.bed) -u \
+| wc -l
+1285
+
+echo "foo" | awk '{print 1285/17680}'
+0.072681</code></pre>
+<ol start="7" style="list-style-type: decimal">
+<li>Create intervals representing the canonical 2bp splice sites on either side of each exon (don’t worry about excluding splice sites at the first or last exon). (Hint - have a look at the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/flank.html">flank</a> tool.)</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools flank -l 2 -r 2 -i exons.bed -g genome.txt > splice-sites.bed</code></pre>
+<p>Or:</p>
+<pre><code>bedtools slop -b 2 -i exons.bed -g genome.txt > exons.plus2.bed
+bedtools subtract -a exons.plus2.bed -b exons.bed > splice-sites.bed</code></pre>
+<ol start="8" style="list-style-type: decimal">
+<li>What is the Jaccard statistic between CpG and hESC enhancers? Compare that to the Jaccard statistic between CpG and hESC promoters. Does the result make sense? (Hint - you will need <code>grep</code>).</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools jaccard -a cpg.bed -b <(grep Enhancer hesc.chromHmm.bed)
+intersection union-intersection jaccard n_intersections
+1148180 132977386 0.0086344 4969
+
+bedtools jaccard -a cpg.bed -b <(grep Promoter hesc.chromHmm.bed)
+intersection union-intersection jaccard n_intersections
+15661111 53551816 0.292448 20402</code></pre>
+<ol start="9" style="list-style-type: decimal">
+<li>What would you expect the Jaccard statistic to look like if promoters were randomly distributed throughout the genome? (Hint - you will need the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/shuffle.html">shuffle</a> tool.)</li>
+</ol>
+<p>Answer:</p>
+<pre><code>bedtools shuffle -i <(grep Promoter hesc.chromHmm.bed) -g genome.txt \
+ | sort -k1,1 -k2,2n \
+> promoters.shuffled.bed
+
+bedtools jaccard -a cpg.bed -b promoters.shuffled.bed
+intersection union-intersection jaccard n_intersections
+294071 68556207 0.00428949 78</code></pre>
+<ol start="10" style="list-style-type: decimal">
+<li>Which hESC ChromHMM state (e.g., 11_Weak_Txn, 10_Txn_Elongation) represents the most number of base pairs in the genome? (Hint: you will need to use <code>awk</code> or <code>perl</code> here, as well as the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/groupby.html">groupby</a> tool.)</li>
+</ol>
+ </div>
+ </div>
+ </div>
+</body>
+</html>
diff --git a/tutorial/answers.md b/tutorial/answers.md
new file mode 100644
index 00000000..95ef89cb
--- /dev/null
+++ b/tutorial/answers.md
@@ -0,0 +1,136 @@
+% bedtools Tutorial
+% Aaron Quinlan
+
+
+Puzzles to help teach you more bedtools.
+========================================
+
+1. Create a BED file representing all of the intervals in the genome
+that are NOT exonic.
+
+Answer:
+
+ bedtools complement -i exons.bed -g genome.txt > notexons.bed
+
+
+2. What is the average distance from GWAS SNPs to the closest exon? (Hint - have a look at the [closest](http://bedtools.readthedocs.org/en/latest/content/tools/closest.html) tool.)
+
+Answer:
+
+ bedtools closest -a gwas.bed -b exons.bed -d | head
+ chr1 1005805 1005806 rs3934834 chr1 1007125 1007955 NM_001205252_exon_0_0_chr1_1007126_r 0 - 1320
+ chr1 1079197 1079198 rs11260603 chr1 1078118 1079434 NR_038869_exon_2_0_chr1_1078119_f 0 + 0
+ chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256456_exon_1_0_chr1_1247398_r 0 - 0
+ chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256460_exon_1_0_chr1_1247398_r 0 - 0
+ chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256462_exon_1_0_chr1_1247398_r 0 - 0
+ chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_001256463_exon_1_0_chr1_1247398_r 0 - 0
+ chr1 1247493 1247494 rs12103 chr1 1247397 1247527 NM_017871_exon_1_0_chr1_1247398_r 0 - 0
+ chr1 2069171 2069172 rs425277 chr1 2066700 2066786 NM_001033581_exon_1_0_chr1_2066701_f 0 + 2386
+ chr1 2069171 2069172 rs425277 chr1 2066700 2066786 NM_001033582_exon_1_0_chr1_2066701_f 0 + 2386
+ chr1 2069171 2069172 rs425277 chr1 2066700 2066786 NM_001242874_exon_1_0_chr1_2066701_f 0 + 2386
+
+ bedtools closest -a gwas.bed -b exons.bed -d \
+ | awk '{ sum += $11 } END { if (NR > 0) print sum / NR }'
+ 46713.1
+
+3. Count how many exons occur in each 500kb interval ("window") in the human genome. (Hint - have a look at the `makewindows` tool.)
+
+Answer:
+
+ bedtools makewindows -g genome.txt -w 500000 > genome.windows.bed
+ bedtools intersect -a genome.windows.bed -b exons.bed -c > genome.windows.exoncount.bedg
+
+or...
+
+ bedtools makewindows -g genome.txt -w 500000 \
+ | bedtools intersect -a - -b exons.bed -c \
+ > genome.windows.exoncount.bedg
+
+4. Are there any exons that are completely overlapped by an enhancer? If so, how many?
+
+Answer:
+
+ bedtools intersect -a exons.bed \
+ -b <(grep Enhancer hesc.chromHmm.bed) \
+ -wa -wb -f 1.0 \
+ | head
+ chr1 948846 948956 NM_005101_exon_0_0_chr1_948847_f 0 + chr1 948337 949337 4_Strong_Enhancer
+ chr1 1051439 1051736 NM_017891_exon_9_0_chr1_1051440_r 0 - chr1 1051337 1051737 6_Weak_Enhancer
+ chr1 1109285 1109306 NM_001130045_exon_0_0_chr1_1109286_f 0 + chr1 1108537 1109537 6_Weak_Enhancer
+ chr1 1109803 1109869 NM_001130045_exon_2_0_chr1_1109804_f 0 + chr1 1109737 1109937 6_Weak_Enhancer
+ chr1 1219357 1219470 NM_001130413_exon_4_0_chr1_1219358_f 0 + chr1 1219137 1220137 7_Weak_Enhancer
+ chr1 1219357 1219470 NR_037668_exon_4_0_chr1_1219358_f 0 + chr1 1219137 1220137 7_Weak_Enhancer
+ chr1 1229202 1229313 NM_030649_exon_1_0_chr1_1229203_r 0 - chr1 1228937 1229937 6_Weak_Enhancer
+ chr1 1229469 1229579 NM_030649_exon_2_0_chr1_1229470_r 0 - chr1 1228937 1229937 6_Weak_Enhancer
+ chr1 1234724 1234736 NM_030649_exon_14_0_chr1_1234725_r 0 - chr1 1234137 1234937 7_Weak_Enhancer
+ chr1 1245060 1245231 NM_153339_exon_4_0_chr1_1245061_f 0 + chr1 1244937 1245337 4_Strong_Enhancer
+
+ bedtools intersect -a exons.bed \
+ -b <(grep Enhancer hesc.chromHmm.bed) \
+ -wa -wb -f 1.0 -u \
+ | wc -l
+ 13746
+
+
+5. What fraction of the GWAS SNPs are exonic?
+
+Answer:
+
+ wc -l gwas.bed
+ 17680 gwas.bed
+
+ bedtools intersect -a gwas.bed -b exons.bed -u | wc -l
+ 1625
+
+ echo "foo" | awk '{print 1625/17680}'
+ 0.0919118
+
+6. What fraction of the GWAS SNPs are lie in either enhancers or promoters in the hESC data we have?
+
+Answer:
+
+ bedtools intersect -a gwas.bed -b <(egrep "Enhancer|Promoter" hesc.chromHmm.bed) -u \
+ | wc -l
+ 1285
+
+ echo "foo" | awk '{print 1285/17680}'
+ 0.072681
+
+7. Create intervals representing the canonical 2bp splice sites on either side of each exon (don't worry about excluding splice sites at the first or last exon). (Hint - have a look at the [flank](http://bedtools.readthedocs.org/en/latest/content/tools/flank.html) tool.)
+
+Answer:
+
+ bedtools flank -l 2 -r 2 -i exons.bed -g genome.txt > splice-sites.bed
+
+Or:
+
+ bedtools slop -b 2 -i exons.bed -g genome.txt > exons.plus2.bed
+ bedtools subtract -a exons.plus2.bed -b exons.bed > splice-sites.bed
+
+
+8. What is the Jaccard statistic between CpG and hESC enhancers? Compare that to the Jaccard statistic between CpG and hESC promoters. Does the result make sense? (Hint - you will need `grep`).
+
+Answer:
+
+ bedtools jaccard -a cpg.bed -b <(grep Enhancer hesc.chromHmm.bed)
+ intersection union-intersection jaccard n_intersections
+ 1148180 132977386 0.0086344 4969
+
+ bedtools jaccard -a cpg.bed -b <(grep Promoter hesc.chromHmm.bed)
+ intersection union-intersection jaccard n_intersections
+ 15661111 53551816 0.292448 20402
+
+
+9. What would you expect the Jaccard statistic to look like if promoters were randomly distributed throughout the genome? (Hint - you will need the [shuffle](http://bedtools.readthedocs.org/en/latest/content/tools/shuffle.html) tool.)
+
+Answer:
+
+ bedtools shuffle -i <(grep Promoter hesc.chromHmm.bed) -g genome.txt \
+ | sort -k1,1 -k2,2n \
+ > promoters.shuffled.bed
+
+ bedtools jaccard -a cpg.bed -b promoters.shuffled.bed
+ intersection union-intersection jaccard n_intersections
+ 294071 68556207 0.00428949 78
+
+10. Which hESC ChromHMM state (e.g., 11_Weak_Txn, 10_Txn_Elongation) represents the most number of base pairs in the genome? (Hint: you will need to use `awk` or `perl` here, as well as the [groupby](http://bedtools.readthedocs.org/en/latest/content/tools/groupby.html) tool.)
diff --git a/tutorial/bedtools.html b/tutorial/bedtools.html
new file mode 100644
index 00000000..9f2c6204
--- /dev/null
+++ b/tutorial/bedtools.html
@@ -0,0 +1,542 @@
+<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
+<html xmlns="http://www.w3.org/1999/xhtml">
+<head>
+ <meta http-equiv="Content-Type" content="text/html; charset=utf-8" />
+ <meta http-equiv="Content-Style-Type" content="text/css" />
+ <meta name="generator" content="pandoc" />
+ <meta name="author" content="Aaron Quinlan" />
+ <title>bedtools Tutorial</title>
+ <style type="text/css">code{white-space: pre;}</style>
+ <link rel="stylesheet" href="bootstrap.css" type="text/css" />
+</head>
+<body>
+ <div class="navbar navbar-static-top">
+ <div class="navbar-inner">
+ <div class="container">
+ <span class="doc-title">bedtools Tutorial</span>
+ <ul class="nav pull-right doc-info">
+ <li><p class="navbar-text">Aaron Quinlan</p></li>
+ </ul>
+ </div>
+ </div>
+ </div>
+ <div class="container">
+ <div class="row">
+ <div class="span12">
+ <h1 id="synopsis">Synopsis</h1>
+<p>Our goal is to work through examples that demonstrate how to explore, process and manipulate genomic interval files (e.g., BED, VCF, BAM) with the <code>bedtools</code> software package.</p>
+<p>Some of our analysis will be based upon the Maurano et al exploration of DnaseI hypersensitivity sites in hundreds of primary tissue types.</p>
+<pre><code>Maurano et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012. Vol. 337 no. 6099 pp. 1190-1195.
+
+www.sciencemag.org/content/337/6099/1190.short</code></pre>
+<p>This tutorial is merely meant as an introduction to whet your appetite. There are many, many more tools and options than presented here. We therefore encourage you to read the bedtools <a href="http://bedtools.readthedocs.org/en/latest/">documentation</a>.</p>
+<div class="alert alert-info" role="alert">
+NOTE: We recommend making your browser window as large as possible because some of the examples yield “wide†results and more screen real estate will help make the results clearer.-
+</div>
+<p><br /></p>
+<h1 id="setup">Setup</h1>
+<p>From the Terminal, create a new directory on your Desktop called <code>bedtools-demo</code> (it doesn’t really matter where you create this directory).</p>
+<pre><code>cd ~/Desktop
+mkdir bedtools-demo</code></pre>
+<p>Navigate into that directory.</p>
+<pre><code>cd bedtools-demo</code></pre>
+<p>Download the sample BED files I have provided.</p>
+<pre><code>curl -O http://quinlanlab.cs.virginia.edu/cshl2013/maurano.dnaseI.tgz
+curl -O http://quinlanlab.cs.virginia.edu/cshl2013/cpg.bed
+curl -O http://quinlanlab.cs.virginia.edu/cshl2013/exons.bed
+curl -O http://quinlanlab.cs.virginia.edu/cshl2013/gwas.bed
+curl -O http://quinlanlab.cs.virginia.edu/cshl2013/genome.txt
+curl -O http://quinlanlab.cs.virginia.edu/cshl2013/hesc.chromHmm.bed</code></pre>
+<p>Now, we need to extract all of the 20 Dnase I hypersensitivity BED files from the “tarball†named <code>maurano.dnaseI.tgz</code>.</p>
+<pre><code>tar -zxvf maurano.dnaseI.tgz
+rm maurano.dnaseI.tgz</code></pre>
+<p>Let’s take a look at what files we now have.</p>
+<pre><code>ls -1</code></pre>
+<p><br /></p>
+<h1 id="what-are-these-files">What are these files?</h1>
+<p>Your directory should now contain 23 BED files and 1 genome file. Twenty of these files (those starting with “f†for “fetal tissueâ€) reflect Dnase I hypersensitivity sites measured in twenty different fetal tissue samples from the brain, heart, intestine, kidney, lung, muscle, skin, and stomach.</p>
+<p>In addition: <code>cpg.bed</code> represents CpG islands in the human genome; <code>exons.bed</code> represents RefSeq exons from human genes; <code>gwas.bed</code> represents human disease-associated SNPs that were identified in genome-wide association studies (GWAS); <code>hesc.chromHmm.bed</code> represents the predicted function (by chromHMM) of each interval in the genome of a human embryonic stem cell based upon ChIP-seq experiments from ENCODE.</p>
+<p>The latter 4 files were extracted from the UCSC Genome Browser’s <a href="http://genome.ucsc.edu/cgi-bin/hgTables?command=start">Table Browser</a>.</p>
+<p>In order to have a rough sense of the data, let’s load the <code>cpg.bed</code>, <code>exons.bed</code>, <code>gwas.bed</code>, and <code>hesc.chromHmm.bed</code> files into <a href="http://www.broadinstitute.org/igv/">IGV</a>. To do this, launch IGV, then click File->Load from File. Then select the four files. IGV will warn you that you need to create an index for a couple of the files. Just click OK, as these indices are created automatically and speed up the processing for IGV.</p>
+<p>Once loaded, navigate to <code>TP53</code> by typing <code>TP53</code> in the search bar. Change the track height to 200 for each track, set the font size to 16 for each track, and change the track colors to match the following image:</p>
+<div class="figure">
+<img src="http://quinlanlab.cs.virginia.edu/cshl2013/igv-tp53.png" />
+</div>
+<p>Visualization in IGV or other browsers such as UCSC is a tremendously useful way to make sure that your results make sense to your eye. Coveniently, a subset of bedtools is built-into IGV!</p>
+<p><br /></p>
+<h1 id="the-bedtools-help">The bedtools help</h1>
+<p>Bedtools is a command-line tool. To bring up the help, just type</p>
+<pre><code>bedtools</code></pre>
+<p>As you can see, there are multiple “subcommands†and for bedtools to work you must tell it which subcommand you want to use. Examples:</p>
+<pre><code>bedtools intersect
+bedtools merge
+bedtools subtract</code></pre>
+<p>What version am I using?</p>
+<pre><code>bedtools --version</code></pre>
+<p>How can I get more help?</p>
+<pre><code>bedtools --contact</code></pre>
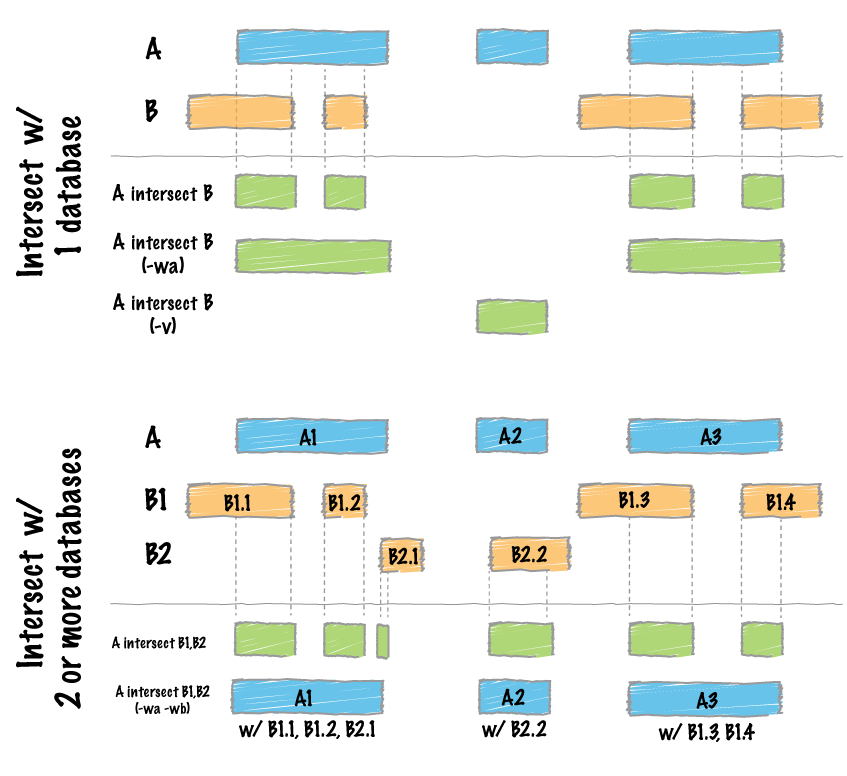
+<h1 id="bedtools-intersect">bedtools “intersectâ€</h1>
+<p>The <code>intersect</code> command is the workhorse of the <code>bedtools</code> suite. It compares two or more BED/BAM/VCF/GFF files and identifies all the regions in the gemome where the features in the two files overlap (that is, share at least one base pair in common).</p>
+<div class="figure">
+<img src="http://bedtools.readthedocs.org/en/latest/_images/intersect-glyph.png" />
+</div>
+<h2 id="default-behavior">Default behavior</h2>
+<p>By default, <code>intersect</code> reports the intervals that represent overlaps between your two files. To demonstrate, let’s identify all of the CpG islands that overlap exons.</p>
+<pre><code>bedtools intersect -a cpg.bed -b exons.bed | head -5
+chr1 29320 29370 CpG:_116
+chr1 135124 135563 CpG:_30
+chr1 327790 328229 CpG:_29
+chr1 327790 328229 CpG:_29
+chr1 327790 328229 CpG:_29</code></pre>
+<div class="alert alert-info" role="alert">
+NOTE: In this case, the intervals reported are NOT the original CpG intervals, but rather a refined interval reflecting solely the portion of each original CpG interval that overlapped with the exon(s).
+</div>
+<h2 id="reporting-the-original-feature-in-each-file.">Reporting the original feature in each file.</h2>
+<p>The <code>-wa</code> (write A) and <code>-wb</code> (write B) options allow one to see the original records from the A and B files that overlapped. As such, instead of not only showing you <em>where</em> the intersections occurred, it shows you <em>what</em> intersected.</p>
+<pre><code>bedtools intersect -a cpg.bed -b exons.bed -wa -wb \
+| head -5
+chr1 28735 29810 CpG:_116 chr1 29320 29370 NR_024540_exon_10_0_chr1_29321_r -
+chr1 135124 135563 CpG:_30 chr1 134772 139696 NR_039983_exon_0_0_chr1_134773_r 0 -
+chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028322_exon_2_0_chr1_324439_f 0 +
+chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028325_exon_2_0_chr1_324439_f 0 +
+chr1 327790 328229 CpG:_29 chr1 327035 328581 NR_028327_exon_3_0_chr1_327036_f 0 +</code></pre>
+<h2 id="how-many-base-pairs-of-overlap-were-there">How many base pairs of overlap were there?</h2>
+<p>The <code>-wo</code> (write overlap) option allows one to also report the <em>number</em> of base pairs of overlap between the features that overlap between each of the files.</p>
+<pre><code>bedtools intersect -a cpg.bed -b exons.bed -wo \
+| head -10
+chr1 28735 29810 CpG:_116 chr1 29320 29370 NR_024540_exon_10_0_chr1_29321_r - 50
+chr1 135124 135563 CpG:_30 chr1 134772 139696 NR_039983_exon_0_0_chr1_134773_r 0 439
+chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028322_exon_2_0_chr1_324439_f 0 439
+chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028325_exon_2_0_chr1_324439_f 0 439
+chr1 327790 328229 CpG:_29 chr1 327035 328581 NR_028327_exon_3_0_chr1_327036_f 0 439
+chr1 713984 714547 CpG:_60 chr1 713663 714068 NR_033908_exon_6_0_chr1_713664_r 0 84
+chr1 762416 763445 CpG:_115 chr1 761585 762902 NR_024321_exon_0_0_chr1_761586_r - 486
+chr1 762416 763445 CpG:_115 chr1 762970 763155 NR_015368_exon_0_0_chr1_762971_f + 185
+chr1 762416 763445 CpG:_115 chr1 762970 763155 NR_047519_exon_0_0_chr1_762971_f + 185
+chr1 762416 763445 CpG:_115 chr1 762970 763155 NR_047520_exon_0_0_chr1_762971_f + 185</code></pre>
+<h2 id="counting-the-number-of-overlapping-features.">Counting the number of overlapping features.</h2>
+<p>We can also count, for each feature in the “A†file, the number of overlapping features in the “B†file. This is handled with the <code>-c</code> option.</p>
+<pre><code>bedtools intersect -a cpg.bed -b exons.bed -c \
+| head
+chr1 28735 29810 CpG:_116 1
+chr1 135124 135563 CpG:_30 1
+chr1 327790 328229 CpG:_29 3
+chr1 437151 438164 CpG:_84 0
+chr1 449273 450544 CpG:_99 0
+chr1 533219 534114 CpG:_94 0
+chr1 544738 546649 CpG:_171 0
+chr1 713984 714547 CpG:_60 1
+chr1 762416 763445 CpG:_115 10
+chr1 788863 789211 CpG:_28 9</code></pre>
+<p><br /></p>
+<h2 id="find-features-that-do-not-overlap">Find features that DO NOT overlap</h2>
+<p>Often we want to identify those features in our A file that <strong>do not</strong> overlap features in the B file. The <code>-v</code> option is your friend in this case.</p>
+<pre><code>bedtools intersect -a cpg.bed -b exons.bed -v \
+| head
+chr1 437151 438164 CpG:_84
+chr1 449273 450544 CpG:_99
+chr1 533219 534114 CpG:_94
+chr1 544738 546649 CpG:_171
+chr1 801975 802338 CpG:_24
+chr1 805198 805628 CpG:_50
+chr1 839694 840619 CpG:_83
+chr1 844299 845883 CpG:_153
+chr1 912869 913153 CpG:_28
+chr1 919726 919927 CpG:_15</code></pre>
+<h2 id="require-a-minimal-fraction-of-overlap.">Require a minimal fraction of overlap.</h2>
+<p>Recall that the default is to report overlaps between features in A and B so long as <em>at least one basepair</em> of overlap exists. However, the <code>-f</code> option allows you to specify what fraction of each feature in A should be overlapped by a feature in B before it is reported.</p>
+<p>Let’s be more strict and require 50% of overlap.</p>
+<pre><code>bedtools intersect -a cpg.bed -b exons.bed \
+-wo -f 0.50 \
+| head
+chr1 135124 135563 CpG:_30 chr1 134772 139696 NR_039983_exon_0_0_chr1_134773_r 0 439
+chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028322_exon_2_0_chr1_324439_f 0 439
+chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028325_exon_2_0_chr1_324439_f 0 439
+chr1 327790 328229 CpG:_29 chr1 327035 328581 NR_028327_exon_3_0_chr1_327036_f 0 439
+chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047519_exon_5_0_chr1_788771_f 0 348
+chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047521_exon_4_0_chr1_788771_f 0 348
+chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047523_exon_3_0_chr1_788771_f 0 348
+chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047524_exon_3_0_chr1_788771_f 0 348
+chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047525_exon_4_0_chr1_788771_f 0 348
+chr1 788863 789211 CpG:_28 chr1 788858 794826 NR_047520_exon_6_0_chr1_788859_f 0 348</code></pre>
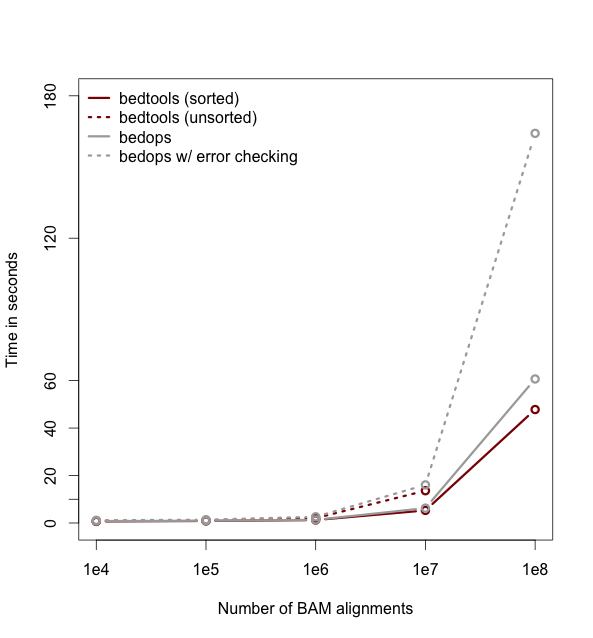
+<h2 id="faster-analysis-via-sorted-data.">Faster analysis via sorted data.</h2>
+<p>So far the examples presented have used the traditional algorithm in bedtools for finding intersections. It turns out, however, that bedtools is much faster when using presorted data.</p>
+<p>For example, compare the difference in speed between the two approaches when finding intersections between <code>exons.bed</code> and <code>hesc.chromHmm.bed</code>:</p>
+<pre><code>time bedtools intersect -a gwas.bed -b hesc.chromHmm.bed > /dev/null
+1.10s user 0.11s system 99% cpu 1.206 total
+
+time bedtools intersect -a gwas.bed -b hesc.chromHmm.bed -sorted > /dev/null
+0.36s user 0.01s system 99% cpu 0.368 total</code></pre>
+<div class="alert alert-info" role="alert">
+NOTE: While the run times in this example are quite small, the performance gains from using the <code>-sorted</code> option groqw as datasets grow larger. For example, compare the runtimes of the sorted and unsorted approaches as a function of dataset size in the figure below. The important thing to remember is that each dataset must be sorted by chromosome and then by start position: <code>sort -k1,1 -k2,2n</code>.-
+</div>
+<div class="figure">
+<img src="http://bedtools.readthedocs.org/en/latest/_images/speed-comparo.png" />
+</div>
+<h2 id="intersecting-multiple-files-at-once.">Intersecting multiple files at once.</h2>
+<p>As of version 2.21.0, bedtools is able to intersect an “A†file against one or more “B†files. This greatly simplifies analyses involving multiple datasets relevant to a given experiment. For example, let’s intersect exons with CpG islands, GWAS SNPs, an the ChromHMM annotations.</p>
+<pre><code>bedtools intersect -a exons.bed -b cpg.bed gwas.bed hesc.chromHmm.bed -sorted | head
+chr1 11873 11937 NR_046018_exon_0_0_chr1_11874_f 0 +
+chr1 11937 12137 NR_046018_exon_0_0_chr1_11874_f 0 +
+chr1 12137 12227 NR_046018_exon_0_0_chr1_11874_f 0 +
+chr1 12612 12721 NR_046018_exon_1_0_chr1_12613_f 0 +
+chr1 13220 14137 NR_046018_exon_2_0_chr1_13221_f 0 +
+chr1 14137 14409 NR_046018_exon_2_0_chr1_13221_f 0 +
+chr1 14361 14829 NR_024540_exon_0_0_chr1_14362_r 0 -
+chr1 14969 15038 NR_024540_exon_1_0_chr1_14970_r 0 -
+chr1 15795 15947 NR_024540_exon_2_0_chr1_15796_r 0 -
+chr1 16606 16765 NR_024540_exon_3_0_chr1_16607_r 0 -</code></pre>
+<p>Now by default, this isn’t incredibly informative as we can’t tell which of the three “B†files yielded the intersection with each exon. However, if we use the <code>-wa</code> and <code>wb</code> options, we can see from which file number (following the order of the files given on the command line) the intersection came. In this case, the 7th column reflects this file number.</p>
+<pre><code>bedtools intersect -a exons.bed -b cpg.bed gwas.bed hesc.chromHmm.bed -sorted -wa -wb \
+ | head -10000 \
+ | tail -10
+chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + 3 chr1 27633213 27635013 5_Strong_Enhancer
+chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + 3 chr1 27635013 27635413 7_Weak_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27632613 27632813 6_Weak_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27632813 27633213 7_Weak_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27633213 27635013 5_Strong_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27635013 27635413 7_Weak_Enhancer
+chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + 1 chr1 27648453 27649006 CpG:_63
+chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + 3 chr1 27648613 27649413 1_Active_Promoter
+chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + 1 chr1 27648453 27649006 CpG:_63
+chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + 3 chr1 27648613 27649413 1_Active_Promoter</code></pre>
+<p>Additionally, one can use file “labels†instead of file numbers to facilitate interpretation, especially when there are <em>many</em> files involved.</p>
+<pre><code>bedtools intersect -a exons.bed -b cpg.bed gwas.bed hesc.chromHmm.bed -sorted -wa -wb -names cpg gwas chromhmm \
+ | head -10000 \
+ | tail -10
+chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27633213 27635013 5_Strong_Enhancer
+chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27635013 27635413 7_Weak_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27632613 27632813 6_Weak_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27632813 27633213 7_Weak_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27633213 27635013 5_Strong_Enhancer
+chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27635013 27635413 7_Weak_Enhancer
+chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + cpg chr1 27648453 27649006 CpG:_63
+chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + chromhmm chr1 27648613 27649413 1_Active_Promoter
+chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + cpg chr1 27648453 27649006 CpG:_63
+chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + chromhmm chr1 27648613 27649413 1_Active_Promoter</code></pre>
+<p><br /></p>
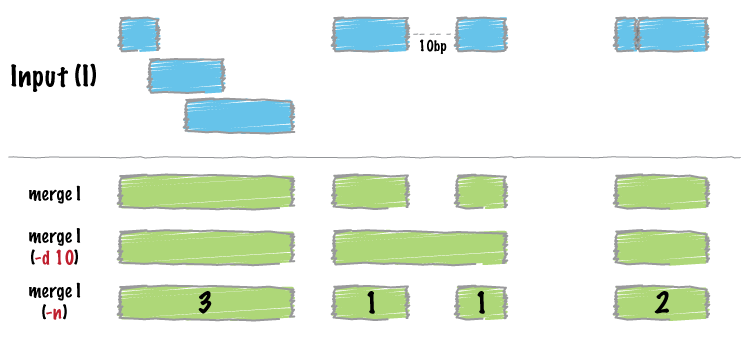
+<h1 id="bedtools-merge">bedtools “mergeâ€</h1>
+<p>Many datasets of genomic features have many individual features that overlap one another (e.g. aligments from a ChiP seq experiment). It is often useful to just cobine the overlapping into a single, contiguous interval. The bedtools <code>merge</code> command will do this for you.</p>
+<div class="figure">
+<img src="http://bedtools.readthedocs.org/en/latest/_images/merge-glyph.png" />
+</div>
+<h2 id="input-must-be-sorted">Input must be sorted</h2>
+<p>The merge tool requires that the input file is sorted by chromosome, then by start position. This allows the merging algorithm to work very quickly without requiring any RAM.</p>
+<p>If your files are unsorted, the <code>merge</code> tool will raise an error. To correct this, you need to sort your BED using the UNIX <code>sort</code> utility. For example:</p>
+<pre><code>sort -k1,1 -k2,2n foo.bed > foo.sort.bed</code></pre>
+<h2 id="merge-intervals.">Merge intervals.</h2>
+<pre><code>Merging results in a new set of intervals representing the merged set of intervals in the input. That is, if a base pair in the genome is covered by 10 features, it will now only be represented once in the output file.
+
+bedtools merge -i exons.bed | head -n 20
+chr1 11873 12227
+chr1 12612 12721
+chr1 13220 14829
+chr1 14969 15038
+chr1 15795 15947
+chr1 16606 16765
+chr1 16857 17055
+chr1 17232 17368
+chr1 17605 17742
+chr1 17914 18061
+chr1 18267 18366
+chr1 24737 24891
+chr1 29320 29370
+chr1 34610 35174
+chr1 35276 35481
+chr1 35720 36081
+chr1 69090 70008
+chr1 134772 139696
+chr1 139789 139847
+chr1 140074 140566</code></pre>
+<h2 id="count-the-number-of-overlapping-intervals.">Count the number of overlapping intervals.</h2>
+<p>A more sophisticated approach would be to not only merge overlapping intervals, but also report the <em>number</em> of intervals that were integrated into the new, merged interval. One does this with the <code>-c</code> and <code>-o</code> options. The <code>-c</code> option allows one to specify a column or columns in the input that you wish to summarize. The <code>-o</code> option defines the operation(s) that you wish to apply to each column listed for the <code>-c</code> option. For example, to count the number of overlapping intervals that led to each of the new “merged†intervals, one will “count†the first column (though the second, third, fourth, etc. would work just fine as well).</p>
+<pre><code>bedtools merge -i exons.bed -c 1 -o count | head -n 20
+chr1 11873 12227 1
+chr1 12612 12721 1
+chr1 13220 14829 2
+chr1 14969 15038 1
+chr1 15795 15947 1
+chr1 16606 16765 1
+chr1 16857 17055 1
+chr1 17232 17368 1
+chr1 17605 17742 1
+chr1 17914 18061 1
+chr1 18267 18366 1
+chr1 24737 24891 1
+chr1 29320 29370 1
+chr1 34610 35174 2
+chr1 35276 35481 2
+chr1 35720 36081 2
+chr1 69090 70008 1
+chr1 134772 139696 1
+chr1 139789 139847 1
+chr1 140074 140566 1</code></pre>
+<h2 id="merging-features-that-are-close-to-one-another.">Merging features that are close to one another.</h2>
+<p>With the <code>-d</code> (distance) option, one can also merge intervals that do not overlap, yet are close to one another. For example, to merge features that are no more than 1000bp apart, one would run:</p>
+<pre><code>bedtools merge -i exons.bed -d 1000 -c 1 -o count | head -20
+chr1 11873 18366 12
+chr1 24737 24891 1
+chr1 29320 29370 1
+chr1 34610 36081 6
+chr1 69090 70008 1
+chr1 134772 140566 3
+chr1 323891 328581 10
+chr1 367658 368597 3
+chr1 621095 622034 3
+chr1 661138 665731 3
+chr1 700244 700627 1
+chr1 701708 701767 1
+chr1 703927 705092 2
+chr1 708355 708487 1
+chr1 709550 709660 1
+chr1 713663 714068 1
+chr1 752750 755214 2
+chr1 761585 763229 10
+chr1 764382 764484 9
+chr1 776579 778984 1</code></pre>
+<h2 id="listing-the-name-of-each-of-the-exons-that-were-merged.">Listing the name of each of the exons that were merged.</h2>
+<p>Many times you want to keep track of the details of exactly which intervals were merged. One way to do this is to create a list of the names of each feature. We can do with with the <code>collapse</code> operation available via the <code>-o</code> argument. The name of the exon is in the fourth column, so we ask <code>merge</code> to create a list of the exon names with <code>-c 4 -o collapse</code>:</p>
+<pre><code>bedtools merge -i exons.bed -d 90 -c 1,4 -o count,collapse | head -20
+chr1 11873 12227 1 NR_046018_exon_0_0_chr1_11874_f
+chr1 12612 12721 1 NR_046018_exon_1_0_chr1_12613_f
+chr1 13220 14829 2 NR_046018_exon_2_0_chr1_13221_f,NR_024540_exon_0_0_chr1_14362_r
+chr1 14969 15038 1 NR_024540_exon_1_0_chr1_14970_r
+chr1 15795 15947 1 NR_024540_exon_2_0_chr1_15796_r
+chr1 16606 16765 1 NR_024540_exon_3_0_chr1_16607_r
+chr1 16857 17055 1 NR_024540_exon_4_0_chr1_16858_r
+chr1 17232 17368 1 NR_024540_exon_5_0_chr1_17233_r
+chr1 17605 17742 1 NR_024540_exon_6_0_chr1_17606_r
+chr1 17914 18061 1 NR_024540_exon_7_0_chr1_17915_r
+chr1 18267 18366 1 NR_024540_exon_8_0_chr1_18268_r
+chr1 24737 24891 1 NR_024540_exon_9_0_chr1_24738_r
+chr1 29320 29370 1 NR_024540_exon_10_0_chr1_29321_r
+chr1 34610 35174 2 NR_026818_exon_0_0_chr1_34611_r,NR_026820_exon_0_0_chr1_34611_r
+chr1 35276 35481 2 NR_026818_exon_1_0_chr1_35277_r,NR_026820_exon_1_0_chr1_35277_r
+chr1 35720 36081 2 NR_026818_exon_2_0_chr1_35721_r,NR_026820_exon_2_0_chr1_35721_r
+chr1 69090 70008 1 NM_001005484_exon_0_0_chr1_69091_f
+chr1 134772 139696 1 NR_039983_exon_0_0_chr1_134773_r
+chr1 139789 139847 1 NR_039983_exon_1_0_chr1_139790_r
+chr1 140074 140566 1 NR_039983_exon_2_0_chr1_140075_r</code></pre>
+<p><br /></p>
+<h1 id="bedtools-complement">bedtools “complementâ€</h1>
+<p>We often want to know which intervals of the genome are <strong>NOT</strong> “covered†by intervals in a given feature file. For example, if you have a set of ChIP-seq peaks, you may also want to know which regions of the genome are not bound by the factor you assayed. The <code>complement</code> addresses this task.</p>
+<div class="figure">
+<img src="http://bedtools.readthedocs.org/en/latest/_images/complement-glyph.png" />
+</div>
+<p>As an example, let’s find all of the non-exonic (i.e., intronic or intergenic) regions of the genome. Note, to do this you need a <a href="http://bedtools.readthedocs.org/en/latest/content/general-usage.html#genome-file-format">“genomeâ€</a> file, which tells <code>bedtools</code> the length of each chromosome in your file. <em>Consider why the tool would need this information…</em></p>
+<pre><code>bedtools complement -i exons.bed -g genome.txt \
+> non-exonic.bed
+head non-exonic.bed
+chr1 0 11873
+chr1 12227 12612
+chr1 12721 13220
+chr1 14829 14969
+chr1 15038 15795
+chr1 15947 16606
+chr1 16765 16857
+chr1 17055 17232
+chr1 17368 17605
+chr1 17742 17914</code></pre>
+<p><br /></p>
+<h1 id="bedtools-genomecov">bedtools “genomecovâ€</h1>
+<p>For many analyses, one wants to measure the genome wide coverage of a feature file. For example, we often want to know what fraction of the genome is covered by 1 feature, 2 features, 3 features, etc. This is frequently crucial when assessing the “uniformity†of coverage from whole-genome sequencing. This is done with the versatile <code>genomecov</code> tool.</p>
+<div class="figure">
+<img src="http://bedtools.readthedocs.org/en/latest/_images/genomecov-glyph.png" />
+</div>
+<p>As an example, let’s produce a histogram of coverage of the exons throughout the genome. Like the <code>merge</code> tool, <code>genomecov</code> requires pre-sorted data. It also needs a genome file as above.</p>
+<pre><code>bedtools genomecov -i exons.bed -g genome.txt</code></pre>
+<p>This should run for 3 minutes or so. At the end of your output, you should see something like:</p>
+<pre><code>genome 0 3062406951 3137161264 0.976171
+genome 1 44120515 3137161264 0.0140638
+genome 2 15076446 3137161264 0.00480576
+genome 3 7294047 3137161264 0.00232505
+genome 4 3650324 3137161264 0.00116358
+genome 5 1926397 3137161264 0.000614057
+genome 6 1182623 3137161264 0.000376972
+genome 7 574102 3137161264 0.000183
+genome 8 353352 3137161264 0.000112634
+genome 9 152653 3137161264 4.86596e-05
+genome 10 113362 3137161264 3.61352e-05
+genome 11 57361 3137161264 1.82844e-05
+genome 12 52000 3137161264 1.65755e-05
+genome 13 55368 3137161264 1.76491e-05
+genome 14 19218 3137161264 6.12592e-06
+genome 15 19369 3137161264 6.17405e-06
+genome 16 26651 3137161264 8.49526e-06
+genome 17 9942 3137161264 3.16911e-06
+genome 18 13442 3137161264 4.28477e-06
+genome 19 1030 3137161264 3.28322e-07
+genome 20 6329 3137161264 2.01743e-06
+...</code></pre>
+<p><br /></p>
+<h2 id="producing-bedgraph-output">Producing BEDGRAPH output</h2>
+<p>Using the <code>-bg</code> option, one can also produce BEDGRAPH output which represents the “depth†fo feature coverage for each base pair in the genome:</p>
+<pre><code>bedtools genomecov -i exons.bed -g genome.txt -bg | head -20
+chr1 11873 12227 1
+chr1 12612 12721 1
+chr1 13220 14361 1
+chr1 14361 14409 2
+chr1 14409 14829 1
+chr1 14969 15038 1
+chr1 15795 15947 1
+chr1 16606 16765 1
+chr1 16857 17055 1
+chr1 17232 17368 1
+chr1 17605 17742 1
+chr1 17914 18061 1
+chr1 18267 18366 1
+chr1 24737 24891 1
+chr1 29320 29370 1
+chr1 34610 35174 2
+chr1 35276 35481 2
+chr1 35720 36081 2
+chr1 69090 70008 1
+chr1 134772 139696 1</code></pre>
+<p><br /></p>
+<h1 id="sophistication-through-chaining-multiple-bedtools">Sophistication through chaining multiple bedtools</h1>
+<p>Analytical power in <code>bedtools</code> comes from the ability to “chain†together multiple tools in order to construct rather sophisicated analyses with very little programming - you just need <strong>genome arithmetic</strong>! Have a look at the examples <a href="http://bedtools.readthedocs.org/en/latest/content/advanced-usage.html">here</a>.</p>
+<p>Here are a few more examples.</p>
+<ol style="list-style-type: decimal">
+<li>Identify the portions of intended capture intervals that did not have any coverage:</li>
+</ol>
+<blockquote class="twitter-tweet" lang="en">
+<p>
+<a href="https://twitter.com/brent_p"><span class="citation">@brent_p</span></a> bedtools genomecov -ibam aln.bam -bga &#10; | awk ‘$4==0’ |
+ | bedtools intersect -a regions -b - > foo
+</p>
+— Aaron Quinlan (<span class="citation">@aaronquinlan</span>) <a href="https://twitter.com/aaronquinlan/status/421786507511205888">January 10, 2014</a>
+</blockquote>
+<script async src="//platform.twitter.com/widgets.js" charset="utf-8"></script>
+<ol start="2" style="list-style-type: decimal">
+<li><a href="http://gettinggeneticsdone.blogspot.com/2014/03/visualize-coverage-exome-targeted-ngs-bedtools.html">Assessing the breadth and depth coverage of sequencing coverage in exome studies</a>.</li>
+</ol>
+<p><br /></p>
+<h1 id="principal-component-analysis">Principal component analysis</h1>
+<p>We will use the bedtools implementation of a Jaccard statistic to meaure the similarity of two datasets. Briefly, the Jaccard statistic measures the ratio of the number of <em>intersecting</em> base pairs to the <em>total</em> number of base pairs in the two sets. As such, the score ranges from 0.0 to 1. 0; lower values reflect lower similarity, whereas higher values reflect higher similarity.</p>
+<p>Let’s walk through an example: we would expect the Dnase hypersensivity sites to be rather similar between two samples of the <strong>same</strong> fetal tissue type. Let’s test:</p>
+<pre><code>bedtools jaccard \
+ -a fHeart-DS16621.hotspot.twopass.fdr0.05.merge.bed \
+ -b fHeart-DS15839.hotspot.twopass.fdr0.05.merge.bed
+intersection union-intersection jaccard n_intersections
+81269248 160493950 0.50637 130852</code></pre>
+<p>But what about the similarity of two <strong>different</strong> tissue types?</p>
+<pre><code>bedtools jaccard \
+ -a fHeart-DS16621.hotspot.twopass.fdr0.05.merge.bed \
+ -b fSkin_fibro_bicep_R-DS19745.hg19.hotspot.twopass.fdr0.05.merge.bed
+intersection union-intersection jaccard n_intersections
+28076951 164197278 0.170995 73261</code></pre>
+<p>Hopefully this demonstrates how the Jaccard statistic can be used as a simple statistic to reduce the dimensionality of the comparison between two large (e.g., often containing thousands or millions of intervals) feature sets.</p>
+<p><br /></p>
+<h2 id="a-jaccard-statistic-for-all-400-pairwise-comparisons.">A Jaccard statistic for all 400 pairwise comparisons.</h2>
+<p>We are going to take this a bit further and use the Jaccard statistic to measure the similarity of all 20 tissue samples against all other 20 samples. Once we have a 20x20 matrix of similarities, we can use dimensionality reduction techniques such as hierarchical clustering or principal component analysis to detect higher order similarities among <strong>all</strong> of the datasets.</p>
+<p>We will use GNU parallel to compute a Jaccard statistic for the 400 (20*20) pairwise comparisons among the fetal tissue samples.</p>
+<p>But first, we need to install <a href="http://www.gnu.org/software/parallel/">GNU parallel</a>.</p>
+<pre><code>brew install parallel</code></pre>
+<p>Next, we need to install a tiny script I wrote for this analysis.</p>
+<pre><code>curl -O http://quinlanlab.cs.virginia.edu/cshl2013/make-matrix.py</code></pre>
+<p>Now, we can use <code>parallel</code> to, you guessed it, compute the 400 pairwise Jaccard statistics in parallel using as many processors as you have available.</p>
+<pre><code>parallel "bedtools jaccard -a {1} -b {2} \
+ | awk 'NR>1' \
+ | cut -f 3 \
+ > {1}.{2}.jaccard" \
+ ::: `ls *.merge.bed` ::: `ls *.merge.bed`</code></pre>
+<p>This command will create a single file containing the pairwise Jaccard measurements from all 400 tests.</p>
+<pre><code>find . \
+ | grep jaccard \
+ | xargs grep "" \
+ | sed -e s"/\.\///" \
+ | perl -pi -e "s/.bed./.bed\t/" \
+ | perl -pi -e "s/.jaccard:/\t/" \
+ > pairwise.dnase.txt</code></pre>
+<p>A bit of cleanup to use more intelligible names for each of the samples.</p>
+<pre><code>cat pairwise.dnase.txt \
+| sed -e 's/.hotspot.twopass.fdr0.05.merge.bed//g' \
+| sed -e 's/.hg19//g' \
+> pairwise.dnase.shortnames.txt</code></pre>
+<p>Now let’s make a 20x20 matrix of the Jaccard statistic. This will allow the data to play nicely with R.</p>
+<pre><code>awk 'NF==3' pairwise.dnase.shortnames.txt \
+| awk '$1 ~ /^f/ && $2 ~ /^f/' \
+| python make-matrix.py \
+> dnase.shortnames.distance.matrix</code></pre>
+<p>Let’s also make a file of labels for each dataset so that we can label each dataset in our R plot.</p>
+<pre><code>cut -f 1 dnase.shortnames.distance.matrix | cut -f 1 -d "-" | cut -f 1 -d "_" > labels.txt</code></pre>
+<p>Now start up R.</p>
+<pre><code>R</code></pre>
+<div class="alert alert-info" role="alert">
+NOTE: The following example assumes that you have both the <code>ggplot2</code> and <code>RColorBrewer</code> packages installed on your computer. If they are not installed, run both <code>install.packages("ggplot2")</code> and <code>install.packages("RColorBrewer")</code> from the R prompt and respond to the prompts that will follow.-
+</div>
+<p>You should see something very similar to this:</p>
+<pre><code>R version 2.15.1 (2012-06-22) -- "Roasted Marshmallows"
+Copyright (C) 2012 The R Foundation for Statistical Computing
+ISBN 3-900051-07-0
+Platform: x86_64-apple-darwin12.0.0 (64-bit)
+
+R is free software and comes with ABSOLUTELY NO WARRANTY.
+You are welcome to redistribute it under certain conditions.
+Type 'license()' or 'licence()' for distribution details.
+
+ Natural language support but running in an English locale
+
+R is a collaborative project with many contributors.
+Type 'contributors()' for more information and
+'citation()' on how to cite R or R packages in publications.
+
+Type 'demo()' for some demos, 'help()' for on-line help, or
+'help.start()' for an HTML browser interface to help.
+Type 'q()' to quit R.
+
+></code></pre>
+<p>No paste these commands into the R console:</p>
+<pre><code>library(ggplot2)
+library(RColorBrewer)
+blues <- colorRampPalette(c('dark blue', 'light blue'))
+greens <- colorRampPalette(c('dark green', 'light green'))
+reds <- colorRampPalette(c('pink', 'dark red'))
+
+setwd("~/Desktop/bedtools-demo")
+x <- read.table('dnase.shortnames.distance.matrix')
+labels <- read.table('labels.txt')
+ngroups <- length(unique(labels))
+pca <- princomp(x)
+qplot(pca$scores[,1], pca$scores[,2], color=labels[,1], geom="point", size=1) +
+ scale_color_manual(values = c(blues(4), greens(5), reds(5))) </code></pre>
+<p>You should see this:</p>
+<div class="figure">
+<img src="http://quinlanlab.cs.virginia.edu/cshl2013/pca.png" />
+</div>
+<p>Et voila.</p>
+<div class="alert alert-info" role="alert">
+Note that PCA was used in this case as a toy example of what PCA does for the CSHL Adv. Seq. course. Heatmaps are a more informative visualization in this case since Jaccard inherently returns a measure of distance.
+</div>
+<p>So let’s make a heatmap for giggles.</p>
+<div class="alert alert-info" role="alert">
+NOTE: The following example assumes that you have both the <code>gplots</code> package installed on your computer. If it are not installed, run <code>install.packages("gplots")</code> from the R prompt and respond to the prompts that will follow.-
+</div>
+<pre><code>library(gplots)
+jaccard_table <- x[, -1]
+jaccard_matrix <- as.matrix(jaccard_table)
+heatmap.2(jaccard_matrix, col = brewer.pal(9,"Blues"), margins = c(14, 14), density.info = "none", lhei=c(2, 8), trace= "none")</code></pre>
+<p>You should see this:</p>
+<div class="figure">
+<img src="http://quinlanlab.cs.virginia.edu/cshl2014/heatmap.png" />
+</div>
+<p><br /></p>
+<h1 id="puzzles-to-help-teach-you-more-bedtools.">Puzzles to help teach you more bedtools.</h1>
+<ol style="list-style-type: decimal">
+<li><p>Create a BED file representing all of the intervals in the genome that are NOT exonic.</p></li>
+<li><p>What is the average distance from GWAS SNPs to the closest exon? (Hint - have a look at the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/closest.html">closest</a> tool.)</p></li>
+<li><p>Count how many exons occur in each 500kb interval (“windowâ€) in the human genome. (Hint - have a look at the <code>makewindows</code> tool.)</p></li>
+<li><p>Are there any exons that are completely overlapped by an enhancer? If so, how many?</p></li>
+<li><p>What fraction of the GWAS SNPs are exonic?</p></li>
+<li><p>What fraction of the GWAS SNPs are lie in either enhancers or promoters in the hESC data we have?</p></li>
+<li><p>Create intervals representing the canonical 2bp splice sites on either side of each exon (don’t worry about excluding splice sites at the first or last exon). (Hint - have a look at the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/flank.html">flank</a> tool.)</p></li>
+<li><p>What is the Jaccard statistic between CpG and hESC enhancers? Compare that to the Jaccard statistic between CpG and hESC promoters. Does the result make sense? (Hint - you will need <code>grep</code>).</p></li>
+<li><p>What would you expect the Jaccard statistic to look like if promoters were randomly distributed throughout the genome? (Hint - you will need the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/shuffle.html">shuffle</a> tool.)</p></li>
+<li><p>Which hESC ChromHMM state (e.g., 11_Weak_Txn, 10_Txn_Elongation) represents the most number of base pairs in the genome? (Hint: you will need to use <code>awk</code> or <code>perl</code> here, as well as the <a href="http://bedtools.readthedocs.org/en/latest/content/tools/groupby.html">groupby</a> tool.)</p></li>
+</ol>
+<p><a href="http://quinlanlab.org/tutorials/cshl2014/answers.html">answers</a></p>
+ </div>
+ </div>
+ </div>
+</body>
+</html>
diff --git a/tutorial/bedtools.md b/tutorial/bedtools.md
new file mode 100644
index 00000000..985afcf7
--- /dev/null
+++ b/tutorial/bedtools.md
@@ -0,0 +1,715 @@
+% bedtools Tutorial
+% Aaron Quinlan
+
+
+Synopsis
+========
+
+Our goal is to work through examples that demonstrate how to
+explore, process and manipulate genomic interval files (e.g., BED, VCF, BAM) with the `bedtools` software package.
+
+Some of our analysis will be based upon the Maurano et al exploration of DnaseI hypersensitivity sites in hundreds of primary tissue types.
+
+ Maurano et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012. Vol. 337 no. 6099 pp. 1190-1195.
+
+ www.sciencemag.org/content/337/6099/1190.short
+
+
+This tutorial is merely meant as an introduction to whet your appetite. There are many, many more tools and options than presented here. We therefore encourage you to read the bedtools [documentation](http://bedtools.readthedocs.org/en/latest/).
+
+
+
+<div class="alert alert-info" role="alert">NOTE: We recommend making your browser window as large as possible because some of the examples yield "wide" results and more screen real estate will help make the results clearer.-</div>
+
+
+\
+
+
+Setup
+=====
+From the Terminal, create a new directory on your Desktop called `bedtools-demo` (it doesn't really matter where you create this directory).
+
+ cd ~/Desktop
+ mkdir bedtools-demo
+
+Navigate into that directory.
+
+ cd bedtools-demo
+
+Download the sample BED files I have provided.
+
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/maurano.dnaseI.tgz
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/cpg.bed
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/exons.bed
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/gwas.bed
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/genome.txt
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/hesc.chromHmm.bed
+
+Now, we need to extract all of the 20 Dnase I hypersensitivity BED files from the "tarball" named
+`maurano.dnaseI.tgz`.
+
+ tar -zxvf maurano.dnaseI.tgz
+ rm maurano.dnaseI.tgz
+
+Let's take a look at what files we now have.
+
+ ls -1
+
+
+\
+
+
+What are these files?
+=========================
+Your directory should now contain 23 BED files and 1 genome file. Twenty of these files (those starting with "f" for "fetal tissue") reflect Dnase I hypersensitivity sites measured in twenty different fetal tissue samples from the brain, heart, intestine, kidney, lung, muscle, skin, and stomach.
+
+In addition: `cpg.bed` represents CpG islands in the human genome; `exons.bed` represents RefSeq exons from human genes; `gwas.bed` represents human disease-associated SNPs that were identified in genome-wide association studies (GWAS); `hesc.chromHmm.bed` represents the predicted function (by chromHMM) of each interval in the genome of a human embryonic stem cell based upon ChIP-seq experiments from ENCODE.
+
+The latter 4 files were extracted from the UCSC Genome Browser's [Table Browser](http://genome.ucsc.edu/cgi-bin/hgTables?command=start).
+
+In order to have a rough sense of the data, let's load the `cpg.bed`, `exons.bed`, `gwas.bed`, and `hesc.chromHmm.bed` files into [IGV](http://www.broadinstitute.org/igv/). To do this, launch IGV, then click File->Load from File. Then select the four files. IGV will warn you that you need to create an index for a couple of the files. Just click OK, as these indices are created automatically and speed up the processing for IGV.
+
+Once loaded, navigate to `TP53` by typing `TP53` in the search bar. Change the track height to 200 for each track, set the font size to 16 for each track, and change the track colors to match the following image:
+
+
+
+Visualization in IGV or other browsers such as UCSC is a tremendously useful way to make sure that your results make sense to your eye. Coveniently, a subset of bedtools is built-into IGV!
+
+
+\
+
+
+The bedtools help
+==================
+Bedtools is a command-line tool. To bring up the help, just type
+
+ bedtools
+
+As you can see, there are multiple "subcommands" and for bedtools to
+work you must tell it which subcommand you want to use. Examples:
+
+ bedtools intersect
+ bedtools merge
+ bedtools subtract
+
+What version am I using?
+
+ bedtools --version
+
+How can I get more help?
+
+ bedtools --contact
+
+
+bedtools "intersect"
+====================
+
+The `intersect` command is the workhorse of the `bedtools` suite. It compares two or more BED/BAM/VCF/GFF files and identifies all the regions in the gemome where the features in the two files overlap (that is, share at least one base pair in common).
+
+
+
+Default behavior
+----------------
+By default, `intersect` reports the intervals that represent overlaps between your two files. To demonstrate, let's identify all of the CpG islands that overlap exons.
+
+ bedtools intersect -a cpg.bed -b exons.bed | head -5
+ chr1 29320 29370 CpG:_116
+ chr1 135124 135563 CpG:_30
+ chr1 327790 328229 CpG:_29
+ chr1 327790 328229 CpG:_29
+ chr1 327790 328229 CpG:_29
+
+<div class="alert alert-info" role="alert">NOTE: In this case, the intervals reported are NOT the original CpG intervals, but rather a refined interval reflecting solely the portion of each original CpG interval that overlapped with the exon(s).</div>
+
+
+Reporting the original feature in each file.
+--------------------------------------------
+The `-wa` (write A) and `-wb` (write B) options allow one to see the original records from the A and B files that overlapped. As such, instead of not only showing you *where* the intersections occurred, it shows you *what* intersected.
+
+ bedtools intersect -a cpg.bed -b exons.bed -wa -wb \
+ | head -5
+ chr1 28735 29810 CpG:_116 chr1 29320 29370 NR_024540_exon_10_0_chr1_29321_r -
+ chr1 135124 135563 CpG:_30 chr1 134772 139696 NR_039983_exon_0_0_chr1_134773_r 0 -
+ chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028322_exon_2_0_chr1_324439_f 0 +
+ chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028325_exon_2_0_chr1_324439_f 0 +
+ chr1 327790 328229 CpG:_29 chr1 327035 328581 NR_028327_exon_3_0_chr1_327036_f 0 +
+
+How many base pairs of overlap were there?
+------------------------------------------
+The `-wo` (write overlap) option allows one to also report the *number* of base pairs of overlap between the features that overlap between each of the files.
+
+ bedtools intersect -a cpg.bed -b exons.bed -wo \
+ | head -10
+ chr1 28735 29810 CpG:_116 chr1 29320 29370 NR_024540_exon_10_0_chr1_29321_r - 50
+ chr1 135124 135563 CpG:_30 chr1 134772 139696 NR_039983_exon_0_0_chr1_134773_r 0 439
+ chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028322_exon_2_0_chr1_324439_f 0 439
+ chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028325_exon_2_0_chr1_324439_f 0 439
+ chr1 327790 328229 CpG:_29 chr1 327035 328581 NR_028327_exon_3_0_chr1_327036_f 0 439
+ chr1 713984 714547 CpG:_60 chr1 713663 714068 NR_033908_exon_6_0_chr1_713664_r 0 84
+ chr1 762416 763445 CpG:_115 chr1 761585 762902 NR_024321_exon_0_0_chr1_761586_r - 486
+ chr1 762416 763445 CpG:_115 chr1 762970 763155 NR_015368_exon_0_0_chr1_762971_f + 185
+ chr1 762416 763445 CpG:_115 chr1 762970 763155 NR_047519_exon_0_0_chr1_762971_f + 185
+ chr1 762416 763445 CpG:_115 chr1 762970 763155 NR_047520_exon_0_0_chr1_762971_f + 185
+
+Counting the number of overlapping features.
+--------------------------------------------
+We can also count, for each feature in the "A" file, the number of overlapping features in the "B" file. This is handled with the `-c` option.
+
+ bedtools intersect -a cpg.bed -b exons.bed -c \
+ | head
+ chr1 28735 29810 CpG:_116 1
+ chr1 135124 135563 CpG:_30 1
+ chr1 327790 328229 CpG:_29 3
+ chr1 437151 438164 CpG:_84 0
+ chr1 449273 450544 CpG:_99 0
+ chr1 533219 534114 CpG:_94 0
+ chr1 544738 546649 CpG:_171 0
+ chr1 713984 714547 CpG:_60 1
+ chr1 762416 763445 CpG:_115 10
+ chr1 788863 789211 CpG:_28 9
+
+\
+
+
+Find features that DO NOT overlap
+--------------------------------------------
+Often we want to identify those features in our A file that **do not** overlap features in the B file. The `-v` option is your friend in this case.
+
+ bedtools intersect -a cpg.bed -b exons.bed -v \
+ | head
+ chr1 437151 438164 CpG:_84
+ chr1 449273 450544 CpG:_99
+ chr1 533219 534114 CpG:_94
+ chr1 544738 546649 CpG:_171
+ chr1 801975 802338 CpG:_24
+ chr1 805198 805628 CpG:_50
+ chr1 839694 840619 CpG:_83
+ chr1 844299 845883 CpG:_153
+ chr1 912869 913153 CpG:_28
+ chr1 919726 919927 CpG:_15
+
+
+Require a minimal fraction of overlap.
+--------------------------------------------
+Recall that the default is to report overlaps between features in A and B so long as *at least one basepair* of overlap exists. However, the `-f` option allows you to specify what fraction of each feature in A should be overlapped by a feature in B before it is reported.
+
+Let's be more strict and require 50% of overlap.
+
+ bedtools intersect -a cpg.bed -b exons.bed \
+ -wo -f 0.50 \
+ | head
+ chr1 135124 135563 CpG:_30 chr1 134772 139696 NR_039983_exon_0_0_chr1_134773_r 0 439
+ chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028322_exon_2_0_chr1_324439_f 0 439
+ chr1 327790 328229 CpG:_29 chr1 324438 328581 NR_028325_exon_2_0_chr1_324439_f 0 439
+ chr1 327790 328229 CpG:_29 chr1 327035 328581 NR_028327_exon_3_0_chr1_327036_f 0 439
+ chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047519_exon_5_0_chr1_788771_f 0 348
+ chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047521_exon_4_0_chr1_788771_f 0 348
+ chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047523_exon_3_0_chr1_788771_f 0 348
+ chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047524_exon_3_0_chr1_788771_f 0 348
+ chr1 788863 789211 CpG:_28 chr1 788770 794826 NR_047525_exon_4_0_chr1_788771_f 0 348
+ chr1 788863 789211 CpG:_28 chr1 788858 794826 NR_047520_exon_6_0_chr1_788859_f 0 348
+
+
+Faster analysis via sorted data.
+--------------------------------------------
+So far the examples presented have used the traditional algorithm in bedtools for finding intersections. It turns out, however, that bedtools is much faster when using presorted data.
+
+For example, compare the difference in speed between the two approaches when finding intersections
+between `exons.bed` and `hesc.chromHmm.bed`:
+
+ time bedtools intersect -a gwas.bed -b hesc.chromHmm.bed > /dev/null
+ 1.10s user 0.11s system 99% cpu 1.206 total
+
+ time bedtools intersect -a gwas.bed -b hesc.chromHmm.bed -sorted > /dev/null
+ 0.36s user 0.01s system 99% cpu 0.368 total
+
+<div class="alert alert-info" role="alert">NOTE: While the run times in this example are quite small, the performance gains from using the `-sorted` option groqw as datasets grow larger. For example, compare the runtimes of the sorted and unsorted approaches as a function of dataset size in the figure below. The important thing to
+remember is that each dataset must be sorted by chromosome and then by start position: `sort -k1,1 -k2,2n`.-</div>
+
+
+
+
+
+Intersecting multiple files at once.
+--------------------------------------------
+As of version 2.21.0, bedtools is able to intersect an "A" file against one or more "B" files. This greatly
+simplifies analyses involving multiple datasets relevant to a given experiment. For example, let's intersect
+exons with CpG islands, GWAS SNPs, an the ChromHMM annotations.
+
+ bedtools intersect -a exons.bed -b cpg.bed gwas.bed hesc.chromHmm.bed -sorted | head
+ chr1 11873 11937 NR_046018_exon_0_0_chr1_11874_f 0 +
+ chr1 11937 12137 NR_046018_exon_0_0_chr1_11874_f 0 +
+ chr1 12137 12227 NR_046018_exon_0_0_chr1_11874_f 0 +
+ chr1 12612 12721 NR_046018_exon_1_0_chr1_12613_f 0 +
+ chr1 13220 14137 NR_046018_exon_2_0_chr1_13221_f 0 +
+ chr1 14137 14409 NR_046018_exon_2_0_chr1_13221_f 0 +
+ chr1 14361 14829 NR_024540_exon_0_0_chr1_14362_r 0 -
+ chr1 14969 15038 NR_024540_exon_1_0_chr1_14970_r 0 -
+ chr1 15795 15947 NR_024540_exon_2_0_chr1_15796_r 0 -
+ chr1 16606 16765 NR_024540_exon_3_0_chr1_16607_r 0 -
+
+Now by default, this isn't incredibly informative as we can't tell which of the three "B" files yielded the intersection with each exon. However, if we use the `-wa` and `wb` options, we can see from which file number (following the order of the files given on the command line) the intersection came. In this case, the 7th column reflects this file number.
+
+ bedtools intersect -a exons.bed -b cpg.bed gwas.bed hesc.chromHmm.bed -sorted -wa -wb \
+ | head -10000 \
+ | tail -10
+ chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + 3 chr1 27633213 27635013 5_Strong_Enhancer
+ chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + 3 chr1 27635013 27635413 7_Weak_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27632613 27632813 6_Weak_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27632813 27633213 7_Weak_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27633213 27635013 5_Strong_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + 3 chr1 27635013 27635413 7_Weak_Enhancer
+ chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + 1 chr1 27648453 27649006 CpG:_63
+ chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + 3 chr1 27648613 27649413 1_Active_Promoter
+ chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + 1 chr1 27648453 27649006 CpG:_63
+ chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + 3 chr1 27648613 27649413 1_Active_Promoter
+
+
+Additionally, one can use file "labels" instead of file numbers to facilitate interpretation, especially when there are _many_ files involved.
+
+ bedtools intersect -a exons.bed -b cpg.bed gwas.bed hesc.chromHmm.bed -sorted -wa -wb -names cpg gwas chromhmm \
+ | head -10000 \
+ | tail -10
+ chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27633213 27635013 5_Strong_Enhancer
+ chr1 27632676 27635124 NM_001276252_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27635013 27635413 7_Weak_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27632613 27632813 6_Weak_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27632813 27633213 7_Weak_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27633213 27635013 5_Strong_Enhancer
+ chr1 27632676 27635124 NM_015023_exon_15_0_chr1_27632677_f 0 + chromhmm chr1 27635013 27635413 7_Weak_Enhancer
+ chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + cpg chr1 27648453 27649006 CpG:_63
+ chr1 27648635 27648882 NM_032125_exon_0_0_chr1_27648636_f 0 + chromhmm chr1 27648613 27649413 1_Active_Promoter
+ chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + cpg chr1 27648453 27649006 CpG:_63
+ chr1 27648635 27648882 NR_037576_exon_0_0_chr1_27648636_f 0 + chromhmm chr1 27648613 27649413 1_Active_Promoter
+
+\
+
+
+bedtools "merge"
+====================
+Many datasets of genomic features have many individual features that overlap one another (e.g. aligments from a ChiP seq experiment). It is often useful to just cobine the overlapping into a single, contiguous interval. The bedtools `merge` command will do this for you.
+
+
+
+
+Input must be sorted
+--------------------
+The merge tool requires that the input file is sorted by chromosome, then by start position. This allows the merging algorithm to work very quickly without requiring any RAM.
+
+If your files are unsorted, the `merge` tool will raise an error. To correct this, you need to sort your BED using the UNIX `sort` utility. For example:
+
+ sort -k1,1 -k2,2n foo.bed > foo.sort.bed
+
+Merge intervals.
+----------------
+
+ Merging results in a new set of intervals representing the merged set of intervals in the input. That is, if a base pair in the genome is covered by 10 features, it will now only be represented once in the output file.
+
+ bedtools merge -i exons.bed | head -n 20
+ chr1 11873 12227
+ chr1 12612 12721
+ chr1 13220 14829
+ chr1 14969 15038
+ chr1 15795 15947
+ chr1 16606 16765
+ chr1 16857 17055
+ chr1 17232 17368
+ chr1 17605 17742
+ chr1 17914 18061
+ chr1 18267 18366
+ chr1 24737 24891
+ chr1 29320 29370
+ chr1 34610 35174
+ chr1 35276 35481
+ chr1 35720 36081
+ chr1 69090 70008
+ chr1 134772 139696
+ chr1 139789 139847
+ chr1 140074 140566
+
+
+
+Count the number of overlapping intervals.
+------------------------------------------
+A more sophisticated approach would be to not only merge overlapping intervals, but also report the *number* of intervals that were integrated into the new, merged interval. One does this with the `-c` and `-o` options. The `-c` option allows one to specify a column or columns in the input that you wish to summarize. The `-o` option defines the operation(s) that you wish to apply to each column listed for the `-c` option. For example, to count the number of overlapping intervals that led to each of the new "merged" intervals, one
+will "count" the first column (though the second, third, fourth, etc. would work just fine as well).
+
+ bedtools merge -i exons.bed -c 1 -o count | head -n 20
+ chr1 11873 12227 1
+ chr1 12612 12721 1
+ chr1 13220 14829 2
+ chr1 14969 15038 1
+ chr1 15795 15947 1
+ chr1 16606 16765 1
+ chr1 16857 17055 1
+ chr1 17232 17368 1
+ chr1 17605 17742 1
+ chr1 17914 18061 1
+ chr1 18267 18366 1
+ chr1 24737 24891 1
+ chr1 29320 29370 1
+ chr1 34610 35174 2
+ chr1 35276 35481 2
+ chr1 35720 36081 2
+ chr1 69090 70008 1
+ chr1 134772 139696 1
+ chr1 139789 139847 1
+ chr1 140074 140566 1
+
+Merging features that are close to one another.
+-----------------------------------------------
+With the `-d` (distance) option, one can also merge intervals that do not overlap, yet are close to one another. For example, to merge features that are no more than 1000bp apart, one would run:
+
+ bedtools merge -i exons.bed -d 1000 -c 1 -o count | head -20
+ chr1 11873 18366 12
+ chr1 24737 24891 1
+ chr1 29320 29370 1
+ chr1 34610 36081 6
+ chr1 69090 70008 1
+ chr1 134772 140566 3
+ chr1 323891 328581 10
+ chr1 367658 368597 3
+ chr1 621095 622034 3
+ chr1 661138 665731 3
+ chr1 700244 700627 1
+ chr1 701708 701767 1
+ chr1 703927 705092 2
+ chr1 708355 708487 1
+ chr1 709550 709660 1
+ chr1 713663 714068 1
+ chr1 752750 755214 2
+ chr1 761585 763229 10
+ chr1 764382 764484 9
+ chr1 776579 778984 1
+
+Listing the name of each of the exons that were merged.
+-------------------------------------------------------
+
+Many times you want to keep track of the details of exactly which intervals were merged. One way to do this is to create a list of the names of each feature. We can do with with the `collapse` operation available via the `-o` argument. The name of the exon is in the fourth column, so we ask `merge` to create a list of the exon names with `-c 4 -o collapse`:
+
+ bedtools merge -i exons.bed -d 90 -c 1,4 -o count,collapse | head -20
+ chr1 11873 12227 1 NR_046018_exon_0_0_chr1_11874_f
+ chr1 12612 12721 1 NR_046018_exon_1_0_chr1_12613_f
+ chr1 13220 14829 2 NR_046018_exon_2_0_chr1_13221_f,NR_024540_exon_0_0_chr1_14362_r
+ chr1 14969 15038 1 NR_024540_exon_1_0_chr1_14970_r
+ chr1 15795 15947 1 NR_024540_exon_2_0_chr1_15796_r
+ chr1 16606 16765 1 NR_024540_exon_3_0_chr1_16607_r
+ chr1 16857 17055 1 NR_024540_exon_4_0_chr1_16858_r
+ chr1 17232 17368 1 NR_024540_exon_5_0_chr1_17233_r
+ chr1 17605 17742 1 NR_024540_exon_6_0_chr1_17606_r
+ chr1 17914 18061 1 NR_024540_exon_7_0_chr1_17915_r
+ chr1 18267 18366 1 NR_024540_exon_8_0_chr1_18268_r
+ chr1 24737 24891 1 NR_024540_exon_9_0_chr1_24738_r
+ chr1 29320 29370 1 NR_024540_exon_10_0_chr1_29321_r
+ chr1 34610 35174 2 NR_026818_exon_0_0_chr1_34611_r,NR_026820_exon_0_0_chr1_34611_r
+ chr1 35276 35481 2 NR_026818_exon_1_0_chr1_35277_r,NR_026820_exon_1_0_chr1_35277_r
+ chr1 35720 36081 2 NR_026818_exon_2_0_chr1_35721_r,NR_026820_exon_2_0_chr1_35721_r
+ chr1 69090 70008 1 NM_001005484_exon_0_0_chr1_69091_f
+ chr1 134772 139696 1 NR_039983_exon_0_0_chr1_134773_r
+ chr1 139789 139847 1 NR_039983_exon_1_0_chr1_139790_r
+ chr1 140074 140566 1 NR_039983_exon_2_0_chr1_140075_r
+
+\
+
+
+bedtools "complement"
+=====================
+We often want to know which intervals of the genome are **NOT** "covered" by intervals in a given feature file. For example, if you have a set of ChIP-seq peaks, you may also want to know which regions of the genome are not bound by the factor you assayed. The `complement` addresses this task.
+
+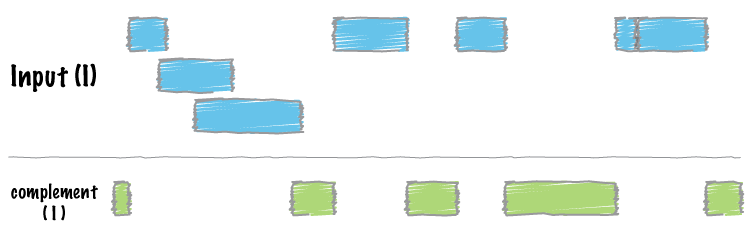
+
+As an example, let's find all of the non-exonic (i.e., intronic or intergenic) regions of the genome. Note, to do this you need a ["genome"](http://bedtools.readthedocs.org/en/latest/content/general-usage.html#genome-file-format) file, which tells `bedtools` the length of each chromosome in your file. *Consider why the tool would need this information...*
+
+ bedtools complement -i exons.bed -g genome.txt \
+ > non-exonic.bed
+ head non-exonic.bed
+ chr1 0 11873
+ chr1 12227 12612
+ chr1 12721 13220
+ chr1 14829 14969
+ chr1 15038 15795
+ chr1 15947 16606
+ chr1 16765 16857
+ chr1 17055 17232
+ chr1 17368 17605
+ chr1 17742 17914
+
+
+\
+
+
+bedtools "genomecov"
+====================
+For many analyses, one wants to measure the genome wide coverage of a feature file. For example, we often want to know what fraction of the genome is covered by 1 feature, 2 features, 3 features, etc. This is frequently crucial when assessing the "uniformity" of coverage from whole-genome sequencing. This is done with the versatile `genomecov` tool.
+
+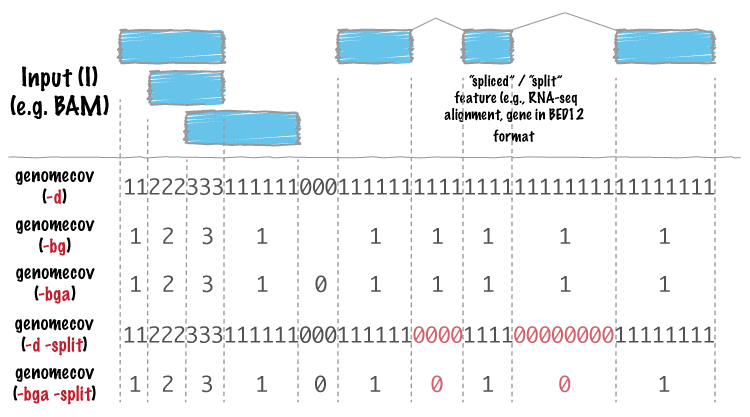
+
+As an example, let's produce a histogram of coverage of the exons throughout the genome. Like the `merge` tool, `genomecov` requires pre-sorted data. It also needs a genome file as above.
+
+ bedtools genomecov -i exons.bed -g genome.txt
+
+This should run for 3 minutes or so. At the end of your output, you should see something like:
+
+ genome 0 3062406951 3137161264 0.976171
+ genome 1 44120515 3137161264 0.0140638
+ genome 2 15076446 3137161264 0.00480576
+ genome 3 7294047 3137161264 0.00232505
+ genome 4 3650324 3137161264 0.00116358
+ genome 5 1926397 3137161264 0.000614057
+ genome 6 1182623 3137161264 0.000376972
+ genome 7 574102 3137161264 0.000183
+ genome 8 353352 3137161264 0.000112634
+ genome 9 152653 3137161264 4.86596e-05
+ genome 10 113362 3137161264 3.61352e-05
+ genome 11 57361 3137161264 1.82844e-05
+ genome 12 52000 3137161264 1.65755e-05
+ genome 13 55368 3137161264 1.76491e-05
+ genome 14 19218 3137161264 6.12592e-06
+ genome 15 19369 3137161264 6.17405e-06
+ genome 16 26651 3137161264 8.49526e-06
+ genome 17 9942 3137161264 3.16911e-06
+ genome 18 13442 3137161264 4.28477e-06
+ genome 19 1030 3137161264 3.28322e-07
+ genome 20 6329 3137161264 2.01743e-06
+ ...
+
+
+\
+
+
+Producing BEDGRAPH output
+--------------------------
+Using the `-bg` option, one can also produce BEDGRAPH output which represents the "depth" fo feature coverage for each base pair in the genome:
+
+ bedtools genomecov -i exons.bed -g genome.txt -bg | head -20
+ chr1 11873 12227 1
+ chr1 12612 12721 1
+ chr1 13220 14361 1
+ chr1 14361 14409 2
+ chr1 14409 14829 1
+ chr1 14969 15038 1
+ chr1 15795 15947 1
+ chr1 16606 16765 1
+ chr1 16857 17055 1
+ chr1 17232 17368 1
+ chr1 17605 17742 1
+ chr1 17914 18061 1
+ chr1 18267 18366 1
+ chr1 24737 24891 1
+ chr1 29320 29370 1
+ chr1 34610 35174 2
+ chr1 35276 35481 2
+ chr1 35720 36081 2
+ chr1 69090 70008 1
+ chr1 134772 139696 1
+
+
+\
+
+
+Sophistication through chaining multiple bedtools
+=================================================
+Analytical power in `bedtools` comes from the ability to "chain" together multiple tools in order to construct rather sophisicated analyses with very little programming - you just need **genome arithmetic**! Have a look at the examples [here](http://bedtools.readthedocs.org/en/latest/content/advanced-usage.html).
+
+Here are a few more examples.
+
+1. Identify the portions of intended capture intervals that did not have any coverage:
+
+<blockquote class="twitter-tweet" lang="en"><p><a href="https://twitter.com/brent_p">@brent_p</a> bedtools genomecov -ibam aln.bam -bga \ | awk '$4==0' | | bedtools intersect -a regions -b - > foo</p>— Aaron Quinlan (@aaronquinlan) <a href="https://twitter.com/aaronquinlan/status/421786507511205888">January 10, 2014</a></blockquote>
+<script async src="//platform.twitter.com/widgets.js" charset="utf-8"></script>
+
+2. [Assessing the breadth and depth coverage of sequencing coverage in exome studies](http://gettinggeneticsdone.blogspot.com/2014/03/visualize-coverage-exome-targeted-ngs-bedtools.html).
+
+\
+
+
+Principal component analysis
+=============================
+
+We will use the bedtools implementation of a Jaccard statistic to meaure the similarity of two
+datasets. Briefly, the Jaccard statistic measures the ratio of the number of *intersecting* base
+pairs to the *total* number of base pairs in the two sets. As such, the score ranges from 0.0 to 1.
+0; lower values reflect lower similarity, whereas higher values reflect higher similarity.
+
+Let's walk through an example: we would expect the Dnase hypersensivity sites to be rather similar
+between two samples of the **same** fetal tissue type. Let's test:
+
+ bedtools jaccard \
+ -a fHeart-DS16621.hotspot.twopass.fdr0.05.merge.bed \
+ -b fHeart-DS15839.hotspot.twopass.fdr0.05.merge.bed
+ intersection union-intersection jaccard n_intersections
+ 81269248 160493950 0.50637 130852
+
+But what about the similarity of two **different** tissue types?
+
+ bedtools jaccard \
+ -a fHeart-DS16621.hotspot.twopass.fdr0.05.merge.bed \
+ -b fSkin_fibro_bicep_R-DS19745.hg19.hotspot.twopass.fdr0.05.merge.bed
+ intersection union-intersection jaccard n_intersections
+ 28076951 164197278 0.170995 73261
+
+Hopefully this demonstrates how the Jaccard statistic can be used as a simple statistic to reduce
+the dimensionality of the comparison between two large (e.g., often containing thousands or
+millions of intervals) feature sets.
+
+
+\
+
+
+A Jaccard statistic for all 400 pairwise comparisons.
+------------------------------------------------------
+
+
+We are going to take this a bit further and use the Jaccard statistic to measure the similarity of
+all 20 tissue samples against all other 20 samples. Once we have a 20x20 matrix of similarities,
+we can use dimensionality reduction techniques such as hierarchical clustering or principal
+component analysis to detect higher order similarities among **all** of the datasets.
+
+
+We will use GNU parallel to compute a Jaccard statistic for the 400 (20*20) pairwise comparisons
+among the fetal tissue samples.
+
+But first, we need to install [GNU parallel](http://www.gnu.org/software/parallel/).
+
+ brew install parallel
+
+Next, we need to install a tiny script I wrote for this analysis.
+
+ curl -O http://quinlanlab.cs.virginia.edu/cshl2013/make-matrix.py
+
+
+Now, we can use `parallel` to, you guessed it, compute the 400 pairwise Jaccard statistics in parallel using as many processors as you have available.
+
+ parallel "bedtools jaccard -a {1} -b {2} \
+ | awk 'NR>1' \
+ | cut -f 3 \
+ > {1}.{2}.jaccard" \
+ ::: `ls *.merge.bed` ::: `ls *.merge.bed`
+
+This command will create a single file containing the pairwise Jaccard measurements from all 400 tests.
+
+ find . \
+ | grep jaccard \
+ | xargs grep "" \
+ | sed -e s"/\.\///" \
+ | perl -pi -e "s/.bed./.bed\t/" \
+ | perl -pi -e "s/.jaccard:/\t/" \
+ > pairwise.dnase.txt
+
+A bit of cleanup to use more intelligible names for each of the samples.
+
+ cat pairwise.dnase.txt \
+ | sed -e 's/.hotspot.twopass.fdr0.05.merge.bed//g' \
+ | sed -e 's/.hg19//g' \
+ > pairwise.dnase.shortnames.txt
+
+Now let's make a 20x20 matrix of the Jaccard statistic. This will allow the data to play nicely with R.
+
+ awk 'NF==3' pairwise.dnase.shortnames.txt \
+ | awk '$1 ~ /^f/ && $2 ~ /^f/' \
+ | python make-matrix.py \
+ > dnase.shortnames.distance.matrix
+
+Let's also make a file of labels for each dataset so that we can label each dataset in our R plot.
+
+ cut -f 1 dnase.shortnames.distance.matrix | cut -f 1 -d "-" | cut -f 1 -d "_" > labels.txt
+
+Now start up R.
+
+ R
+
+<div class="alert alert-info" role="alert">NOTE: The following example assumes that you have both the `ggplot2` and `RColorBrewer` packages installed on your computer. If they
+ are not installed, run both `install.packages("ggplot2")` and `install.packages("RColorBrewer")` from the R prompt and respond to the prompts that will follow.-</div>
+
+You should see something very similar to this:
+
+
+ R version 2.15.1 (2012-06-22) -- "Roasted Marshmallows"
+ Copyright (C) 2012 The R Foundation for Statistical Computing
+ ISBN 3-900051-07-0
+ Platform: x86_64-apple-darwin12.0.0 (64-bit)
+
+ R is free software and comes with ABSOLUTELY NO WARRANTY.
+ You are welcome to redistribute it under certain conditions.
+ Type 'license()' or 'licence()' for distribution details.
+
+ Natural language support but running in an English locale
+
+ R is a collaborative project with many contributors.
+ Type 'contributors()' for more information and
+ 'citation()' on how to cite R or R packages in publications.
+
+ Type 'demo()' for some demos, 'help()' for on-line help, or
+ 'help.start()' for an HTML browser interface to help.
+ Type 'q()' to quit R.
+
+ >
+
+No paste these commands into the R console:
+
+ library(ggplot2)
+ library(RColorBrewer)
+ blues <- colorRampPalette(c('dark blue', 'light blue'))
+ greens <- colorRampPalette(c('dark green', 'light green'))
+ reds <- colorRampPalette(c('pink', 'dark red'))
+
+ setwd("~/Desktop/bedtools-demo")
+ x <- read.table('dnase.shortnames.distance.matrix')
+ labels <- read.table('labels.txt')
+ ngroups <- length(unique(labels))
+ pca <- princomp(x)
+ qplot(pca$scores[,1], pca$scores[,2], color=labels[,1], geom="point", size=1) +
+ scale_color_manual(values = c(blues(4), greens(5), reds(5)))
+
+You should see this:
+
+
+
+Et voila.
+
+<div class="alert alert-info" role="alert">Note that PCA was used in this case as a toy example of what PCA does for the CSHL Adv. Seq. course. Heatmaps are a more informative visualization in this case since Jaccard inherently returns a measure of distance.</div>
+
+
+So let's make a heatmap for giggles.
+
+<div class="alert alert-info" role="alert">NOTE: The following example assumes that you have both the `gplots` package installed on your computer. If it
+ are not installed, run `install.packages("gplots")` from the R prompt and respond to the prompts that will follow.-</div>
+
+ library(gplots)
+ jaccard_table <- x[, -1]
+ jaccard_matrix <- as.matrix(jaccard_table)
+ heatmap.2(jaccard_matrix, col = brewer.pal(9,"Blues"), margins = c(14, 14), density.info = "none", lhei=c(2, 8), trace= "none")
+
+You should see this:
+
+
+
+\
+
+
+Puzzles to help teach you more bedtools.
+========================================
+
+
+1. Create a BED file representing all of the intervals in the genome
+that are NOT exonic.
+
+2. What is the average distance from GWAS SNPs to the closest exon? (Hint - have a look at the [closest](http://bedtools.readthedocs.org/en/latest/content/tools/closest.html) tool.)
+
+3. Count how many exons occur in each 500kb interval ("window") in the human genome. (Hint - have a look at the `makewindows` tool.)
+
+4. Are there any exons that are completely overlapped by an enhancer? If so, how many?
+
+5. What fraction of the GWAS SNPs are exonic?
+
+6. What fraction of the GWAS SNPs are lie in either enhancers or promoters in the hESC data we have?
+
+7. Create intervals representing the canonical 2bp splice sites on either side of each exon (don't worry about excluding splice sites at the first or last exon). (Hint - have a look at the [flank](http://bedtools.readthedocs.org/en/latest/content/tools/flank.html) tool.)
+
+8. What is the Jaccard statistic between CpG and hESC enhancers? Compare that to the Jaccard statistic between CpG and hESC promoters. Does the result make sense? (Hint - you will need `grep`).
+
+9. What would you expect the Jaccard statistic to look like if promoters were randomly distributed throughout the genome? (Hint - you will need the [shuffle](http://bedtools.readthedocs.org/en/latest/content/tools/shuffle.html) tool.)
+
+10. Which hESC ChromHMM state (e.g., 11_Weak_Txn, 10_Txn_Elongation) represents the most number of base pairs in the genome? (Hint: you will need to use `awk` or `perl` here, as well as the [groupby](http://bedtools.readthedocs.org/en/latest/content/tools/groupby.html) tool.)
+
+
+[answers](http://quinlanlab.org/tutorials/cshl2014/answers.html)
diff --git a/tutorial/bootstrap.css b/tutorial/bootstrap.css
new file mode 100644
index 00000000..c1f05c38
--- /dev/null
+++ b/tutorial/bootstrap.css
@@ -0,0 +1,4480 @@
+/*!
+ * Bootstrap v2.0.4
+ *
+ * Copyright 2012 Twitter, Inc
+ * Licensed under the Apache License v2.0
+ * http://www.apache.org/licenses/LICENSE-2.0
+ *
+ * Designed and built with all the love in the world @twitter by @mdo and @fat.
+ */
+@import url(https://fonts.googleapis.com/css?family=Lato:300italic,700italic,300,700)
+.clearfix {
+ *zoom: 1;
+}
+.clearfix:before,
+.clearfix:after {
+ display: table;
+ content: "";
+}
+.clearfix:after {
+ clear: both;
+}
+.hide-text {
+ font: 0/0 a;
+ color: transparent;
+ text-shadow: none;
+ background-color: transparent;
+ border: 0;
+}
+.input-block-level {
+ display: block;
+ width: 100%;
+ min-height: 28px;
+ -webkit-box-sizing: border-box;
+ -moz-box-sizing: border-box;
+ -ms-box-sizing: border-box;
+ box-sizing: border-box;
+}
+article,
+aside,
+details,
+figcaption,
+figure,
+footer,
+header,
+hgroup,
+nav,
+section {
+ display: block;
+}
+audio,
+canvas,
+video {
+ display: inline-block;
+ *display: inline;
+ *zoom: 1;
+}
+audio:not([controls]) {
+ display: none;
+}
+html {
+ font-size: 100%;
+ -webkit-text-size-adjust: 100%;
+ -ms-text-size-adjust: 100%;
+}
+a:focus {
+ outline: thin dotted #333;
+ outline: 5px auto -webkit-focus-ring-color;
+ outline-offset: -2px;
+}
+a:hover,
+a:active {
+ outline: 0;
+}
+sub,
+sup {
+ position: relative;
+ font-size: 75%;
+ line-height: 0;
+ vertical-align: baseline;
+}
+sup {
+ top: -0.5em;
+}
+sub {
+ bottom: -0.25em;
+}
+img {
+ max-width: 100%;
+ vertical-align: middle;
+ border: 0;
+ -ms-interpolation-mode: bicubic;
+}
+#map_canvas img {
+ max-width: none;
+}
+button,
+input,
+select,
+textarea {
+ margin: 0;
+ font-size: 100%;
+ vertical-align: middle;
+}
+button,
+input {
+ *overflow: visible;
+ line-height: normal;
+}
+button::-moz-focus-inner,
+input::-moz-focus-inner {
+ padding: 0;
+ border: 0;
+}
+button,
+input[type="button"],
+input[type="reset"],
+input[type="submit"] {
+ cursor: pointer;
+ -webkit-appearance: button;
+}
+input[type="search"] {
+ -webkit-box-sizing: content-box;
+ -moz-box-sizing: content-box;
+ box-sizing: content-box;
+ -webkit-appearance: textfield;
+}
+input[type="search"]::-webkit-search-decoration,
+input[type="search"]::-webkit-search-cancel-button {
+ -webkit-appearance: none;
+}
+textarea {
+ overflow: auto;
+ vertical-align: top;
+}
+body {
+ margin: 0;
+/* font-family: "Helvetica Neue Light", Helvetica, Arial, sans-serif; */
+ font-family: "Helvetica Neue", Helvetica, Arial, sans-serif;
+
+/* font-family: Century Gothic, sans-serif;*/
+/* font-family: "Lucida Sans Unicode", "Lucida Grande", sans-serif;*/
+ font-size: 16px;
+ line-height: 24px;
+ color: #333333;
+ background-color: #ffffff;
+}
+a {
+ color: #3a87ad;
+ text-decoration: none;
+}
+a:hover {
+ color: #005580;
+ text-decoration: underline;
+}
+.row {
+ margin-left: -20px;
+ *zoom: 1;
+}
+.row:before,
+.row:after {
+ display: table;
+ content: "";
+}
+.row:after {
+ clear: both;
+}
+[class*="span"] {
+ float: left;
+ margin-left: 20px;
+}
+.container,
+.navbar-fixed-top .container,
+.navbar-fixed-bottom .container {
+ width: 940px;
+}
+.span12 {
+ width: 940px;
+}
+.span11 {
+ width: 860px;
+}
+.span10 {
+ width: 780px;
+}
+.span9 {
+ width: 700px;
+}
+.span8 {
+ width: 620px;
+}
+.span7 {
+ width: 540px;
+}
+.span6 {
+ width: 460px;
+}
+.span5 {
+ width: 380px;
+}
+.span4 {
+ width: 300px;
+}
+.span3 {
+ width: 220px;
+}
+.span2 {
+ width: 140px;
+}
+.span1 {
+ width: 60px;
+}
+.offset12 {
+ margin-left: 980px;
+}
+.offset11 {
+ margin-left: 900px;
+}
+.offset10 {
+ margin-left: 820px;
+}
+.offset9 {
+ margin-left: 740px;
+}
+.offset8 {
+ margin-left: 660px;
+}
+.offset7 {
+ margin-left: 580px;
+}
+.offset6 {
+ margin-left: 500px;
+}
+.offset5 {
+ margin-left: 420px;
+}
+.offset4 {
+ margin-left: 340px;
+}
+.offset3 {
+ margin-left: 260px;
+}
+.offset2 {
+ margin-left: 180px;
+}
+.offset1 {
+ margin-left: 100px;
+}
+.row-fluid {
+ width: 100%;
+ *zoom: 1;
+}
+.row-fluid:before,
+.row-fluid:after {
+ display: table;
+ content: "";
+}
+.row-fluid:after {
+ clear: both;
+}
+.row-fluid [class*="span"] {
+ display: block;
+ width: 100%;
+ min-height: 28px;
+ -webkit-box-sizing: border-box;
+ -moz-box-sizing: border-box;
+ -ms-box-sizing: border-box;
+ box-sizing: border-box;
+ float: left;
+ margin-left: 2.127659574%;
+ *margin-left: 2.0744680846382977%;
+}
+.row-fluid [class*="span"]:first-child {
+ margin-left: 0;
+}
+.row-fluid .span12 {
+ width: 99.99999998999999%;
+ *width: 99.94680850063828%;
+}
+.row-fluid .span11 {
+ width: 91.489361693%;
+ *width: 91.4361702036383%;
+}
+.row-fluid .span10 {
+ width: 82.97872339599999%;
+ *width: 82.92553190663828%;
+}
+.row-fluid .span9 {
+ width: 74.468085099%;
+ *width: 74.4148936096383%;
+}
+.row-fluid .span8 {
+ width: 65.95744680199999%;
+ *width: 65.90425531263828%;
+}
+.row-fluid .span7 {
+ width: 57.446808505%;
+ *width: 57.3936170156383%;
+}
+.row-fluid .span6 {
+ width: 48.93617020799999%;
+ *width: 48.88297871863829%;
+}
+.row-fluid .span5 {
+ width: 40.425531911%;
+ *width: 40.3723404216383%;
+}
+.row-fluid .span4 {
+ width: 31.914893614%;
+ *width: 31.8617021246383%;
+}
+.row-fluid .span3 {
+ width: 23.404255317%;
+ *width: 23.3510638276383%;
+}
+.row-fluid .span2 {
+ width: 14.89361702%;
+ *width: 14.8404255306383%;
+}
+.row-fluid .span1 {
+ width: 6.382978723%;
+ *width: 6.329787233638298%;
+}
+.container {
+ margin-right: auto;
+ margin-left: auto;
+ *zoom: 1;
+}
+.container:before,
+.container:after {
+ display: table;
+ content: "";
+}
+.container:after {
+ clear: both;
+}
+.container-fluid {
+ padding-right: 20px;
+ padding-left: 20px;
+ *zoom: 1;
+}
+.container-fluid:before,
+.container-fluid:after {
+ display: table;
+ content: "";
+}
+.container-fluid:after {
+ clear: both;
+}
+p {
+ margin: 0 0 9px;
+}
+p small {
+ font-size: 11px;
+ color: #999999;
+}
+.lead {
+ margin-bottom: 18px;
+ font-size: 20px;
+ font-weight: 200;
+ line-height: 27px;
+}
+h1,
+h2,
+h3,
+h4,
+h5,
+h6 {
+ margin: 0;
+ font-family: inherit;
+ font-weight: bold;
+ color: inherit;
+ text-rendering: optimizelegibility;
+}
+h1 small,
+h2 small,
+h3 small,
+h4 small,
+h5 small,
+h6 small {
+ font-weight: normal;
+ color: #999999;
+}
+h1 {
+ font-size: 30px;
+ line-height: 36px;
+}
+h1 small {
+ font-size: 18px;
+}
+h2 {
+ font-size: 24px;
+ line-height: 36px;
+}
+h2 small {
+ font-size: 18px;
+}
+h3 {
+ font-size: 18px;
+ line-height: 27px;
+}
+h3 small {
+ font-size: 14px;
+}
+h4,
+h5,
+h6 {
+ line-height: 18px;
+}
+h4 {
+ font-size: 14px;
+}
+h4 small {
+ font-size: 12px;
+}
+h5 {
+ font-size: 12px;
+}
+h6 {
+ font-size: 11px;
+ color: #999999;
+ text-transform: uppercase;
+}
+.page-header {
+ padding-bottom: 17px;
+ margin: 18px 0;
+ border-bottom: 1px solid #eeeeee;
+}
+.page-header h1 {
+ line-height: 1;
+}
+ul,
+ol {
+ padding: 0;
+ margin: 0 0 9px 25px;
+}
+ul ul,
+ul ol,
+ol ol,
+ol ul {
+ margin-bottom: 0;
+}
+ul {
+ list-style: disc;
+}
+ol {
+ list-style: decimal;
+}
+li {
+ line-height: 18px;
+}
+ul.unstyled,
+ol.unstyled {
+ margin-left: 0;
+ list-style: none;
+}
+dl {
+ margin-bottom: 18px;
+}
+dt,
+dd {
+ line-height: 18px;
+}
+dt {
+ font-weight: bold;
+ line-height: 17px;
+}
+dd {
+ margin-left: 9px;
+}
+.dl-horizontal dt {
+ float: left;
+ width: 120px;
+ clear: left;
+ text-align: right;
+ overflow: hidden;
+ text-overflow: ellipsis;
+ white-space: nowrap;
+}
+.dl-horizontal dd {
+ margin-left: 130px;
+}
+hr {
+ margin: 18px 0;
+ border: 0;
+ border-top: 1px solid #eeeeee;
+ border-bottom: 1px solid #ffffff;
+}
+strong {
+ font-weight: bold;
+}
+em {
+ font-style: italic;
+}
+.muted {
+ color: #999999;
+}
+abbr[title] {
+ cursor: help;
+ border-bottom: 1px dotted #999999;
+}
+abbr.initialism {
+ font-size: 90%;
+ text-transform: uppercase;
+}
+blockquote {
+ padding: 0 0 0 15px;
+ margin: 0 0 18px;
+ border-left: 5px solid #eeeeee;
+}
+blockquote p {
+ margin-bottom: 0;
+ font-size: 16px;
+ font-weight: 300;
+ line-height: 22.5px;
+}
+blockquote small {
+ display: block;
+ line-height: 18px;
+ color: #999999;
+}
+blockquote small:before {
+ content: '\2014 \00A0';
+}
+blockquote.pull-right {
+ float: right;
+ padding-right: 15px;
+ padding-left: 0;
+ border-right: 5px solid #eeeeee;
+ border-left: 0;
+}
+blockquote.pull-right p,
+blockquote.pull-right small {
+ text-align: right;
+}
+q:before,
+q:after,
+blockquote:before,
+blockquote:after {
+ content: "";
+}
+address {
+ display: block;
+ margin-bottom: 18px;
+ font-style: normal;
+ line-height: 18px;
+}
+small {
+ font-size: 100%;
+}
+cite {
+ font-style: normal;
+}
+code,
+pre {
+ padding: 0 3px 2px;
+ font-family: Menlo, Monaco, Consolas, "Courier New", monospace;
+ font-size: 12px;
+ color: #333333;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+}
+code {
+ padding: 2px 4px;
+ color: #d14;
+ background-color: #f7f7f9;
+ border: 1px solid #e1e1e8;
+}
+pre {
+ display: block;
+ padding: 8.5px;
+ margin: 0 0 9px;
+ font-size: 12.025px;
+ line-height: 18px;
+ word-break: break-all;
+ word-wrap: break-word;
+ white-space: pre;
+ white-space: pre-wrap;
+ background-color: #f5f5f5;
+ border: 1px solid #ccc;
+ border: 1px solid rgba(0, 0, 0, 0.15);
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+pre.prettyprint {
+ margin-bottom: 18px;
+}
+pre code {
+ padding: 0;
+ color: inherit;
+ background-color: transparent;
+ border: 0;
+}
+.pre-scrollable {
+ max-height: 340px;
+ overflow-y: scroll;
+}
+.label,
+.badge {
+ font-size: 10.998px;
+ font-weight: bold;
+ line-height: 14px;
+ color: #ffffff;
+ vertical-align: baseline;
+ white-space: nowrap;
+ text-shadow: 0 -1px 0 rgba(0, 0, 0, 0.25);
+ background-color: #999999;
+}
+.label {
+ padding: 1px 4px 2px;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+}
+.badge {
+ padding: 1px 9px 2px;
+ -webkit-border-radius: 9px;
+ -moz-border-radius: 9px;
+ border-radius: 9px;
+}
+a.label:hover,
+a.badge:hover {
+ color: #ffffff;
+ text-decoration: none;
+ cursor: pointer;
+}
+.label-important,
+.badge-important {
+ background-color: #b94a48;
+}
+.label-important[href],
+.badge-important[href] {
+ background-color: #953b39;
+}
+.label-warning,
+.badge-warning {
+ background-color: #f89406;
+}
+.label-warning[href],
+.badge-warning[href] {
+ background-color: #c67605;
+}
+.label-success,
+.badge-success {
+ background-color: #468847;
+}
+.label-success[href],
+.badge-success[href] {
+ background-color: #356635;
+}
+.label-info,
+.badge-info {
+ background-color: #3a87ad;
+}
+.label-info[href],
+.badge-info[href] {
+ background-color: #2d6987;
+}
+.label-inverse,
+.badge-inverse {
+ background-color: #333333;
+}
+.label-inverse[href],
+.badge-inverse[href] {
+ background-color: #1a1a1a;
+}
+table {
+ max-width: 100%;
+ background-color: transparent;
+ border-collapse: collapse;
+ border-spacing: 0;
+}
+.table {
+ width: 100%;
+ margin-bottom: 18px;
+}
+.table th,
+.table td {
+ padding: 8px;
+ line-height: 18px;
+ text-align: left;
+ vertical-align: top;
+ border-top: 1px solid #dddddd;
+}
+.table th {
+ font-weight: bold;
+}
+.table thead th {
+ vertical-align: bottom;
+}
+.table caption + thead tr:first-child th,
+.table caption + thead tr:first-child td,
+.table colgroup + thead tr:first-child th,
+.table colgroup + thead tr:first-child td,
+.table thead:first-child tr:first-child th,
+.table thead:first-child tr:first-child td {
+ border-top: 0;
+}
+.table tbody + tbody {
+ border-top: 2px solid #dddddd;
+}
+.table-condensed th,
+.table-condensed td {
+ padding: 4px 5px;
+}
+.table-bordered {
+ border: 1px solid #dddddd;
+ border-collapse: separate;
+ *border-collapse: collapsed;
+ border-left: 0;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+.table-bordered th,
+.table-bordered td {
+ border-left: 1px solid #dddddd;
+}
+.table-bordered caption + thead tr:first-child th,
+.table-bordered caption + tbody tr:first-child th,
+.table-bordered caption + tbody tr:first-child td,
+.table-bordered colgroup + thead tr:first-child th,
+.table-bordered colgroup + tbody tr:first-child th,
+.table-bordered colgroup + tbody tr:first-child td,
+.table-bordered thead:first-child tr:first-child th,
+.table-bordered tbody:first-child tr:first-child th,
+.table-bordered tbody:first-child tr:first-child td {
+ border-top: 0;
+}
+.table-bordered thead:first-child tr:first-child th:first-child,
+.table-bordered tbody:first-child tr:first-child td:first-child {
+ -webkit-border-top-left-radius: 4px;
+ border-top-left-radius: 4px;
+ -moz-border-radius-topleft: 4px;
+}
+.table-bordered thead:first-child tr:first-child th:last-child,
+.table-bordered tbody:first-child tr:first-child td:last-child {
+ -webkit-border-top-right-radius: 4px;
+ border-top-right-radius: 4px;
+ -moz-border-radius-topright: 4px;
+}
+.table-bordered thead:last-child tr:last-child th:first-child,
+.table-bordered tbody:last-child tr:last-child td:first-child {
+ -webkit-border-radius: 0 0 0 4px;
+ -moz-border-radius: 0 0 0 4px;
+ border-radius: 0 0 0 4px;
+ -webkit-border-bottom-left-radius: 4px;
+ border-bottom-left-radius: 4px;
+ -moz-border-radius-bottomleft: 4px;
+}
+.table-bordered thead:last-child tr:last-child th:last-child,
+.table-bordered tbody:last-child tr:last-child td:last-child {
+ -webkit-border-bottom-right-radius: 4px;
+ border-bottom-right-radius: 4px;
+ -moz-border-radius-bottomright: 4px;
+}
+.table-striped tbody tr:nth-child(odd) td,
+.table-striped tbody tr:nth-child(odd) th {
+ background-color: #f9f9f9;
+}
+.table tbody tr:hover td,
+.table tbody tr:hover th {
+ background-color: #f5f5f5;
+}
+table .span1 {
+ float: none;
+ width: 44px;
+ margin-left: 0;
+}
+table .span2 {
+ float: none;
+ width: 124px;
+ margin-left: 0;
+}
+table .span3 {
+ float: none;
+ width: 204px;
+ margin-left: 0;
+}
+table .span4 {
+ float: none;
+ width: 284px;
+ margin-left: 0;
+}
+table .span5 {
+ float: none;
+ width: 364px;
+ margin-left: 0;
+}
+table .span6 {
+ float: none;
+ width: 444px;
+ margin-left: 0;
+}
+table .span7 {
+ float: none;
+ width: 524px;
+ margin-left: 0;
+}
+table .span8 {
+ float: none;
+ width: 604px;
+ margin-left: 0;
+}
+table .span9 {
+ float: none;
+ width: 684px;
+ margin-left: 0;
+}
+table .span10 {
+ float: none;
+ width: 764px;
+ margin-left: 0;
+}
+table .span11 {
+ float: none;
+ width: 844px;
+ margin-left: 0;
+}
+table .span12 {
+ float: none;
+ width: 924px;
+ margin-left: 0;
+}
+table .span13 {
+ float: none;
+ width: 1004px;
+ margin-left: 0;
+}
+table .span14 {
+ float: none;
+ width: 1084px;
+ margin-left: 0;
+}
+table .span15 {
+ float: none;
+ width: 1164px;
+ margin-left: 0;
+}
+table .span16 {
+ float: none;
+ width: 1244px;
+ margin-left: 0;
+}
+table .span17 {
+ float: none;
+ width: 1324px;
+ margin-left: 0;
+}
+table .span18 {
+ float: none;
+ width: 1404px;
+ margin-left: 0;
+}
+table .span19 {
+ float: none;
+ width: 1484px;
+ margin-left: 0;
+}
+table .span20 {
+ float: none;
+ width: 1564px;
+ margin-left: 0;
+}
+table .span21 {
+ float: none;
+ width: 1644px;
+ margin-left: 0;
+}
+table .span22 {
+ float: none;
+ width: 1724px;
+ margin-left: 0;
+}
+table .span23 {
+ float: none;
+ width: 1804px;
+ margin-left: 0;
+}
+table .span24 {
+ float: none;
+ width: 1884px;
+ margin-left: 0;
+}
+form {
+ margin: 0 0 18px;
+}
+fieldset {
+ padding: 0;
+ margin: 0;
+ border: 0;
+}
+legend {
+ display: block;
+ width: 100%;
+ padding: 0;
+ margin-bottom: 27px;
+ font-size: 19.5px;
+ line-height: 36px;
+ color: #333333;
+ border: 0;
+ border-bottom: 1px solid #e5e5e5;
+}
+legend small {
+ font-size: 13.5px;
+ color: #999999;
+}
+label,
+input,
+button,
+select,
+textarea {
+ font-size: 13px;
+ font-weight: normal;
+ line-height: 18px;
+}
+input,
+button,
+select,
+textarea {
+ font-family: "Helvetica Neue", Helvetica, Arial, sans-serif;
+}
+label {
+ display: block;
+ margin-bottom: 5px;
+}
+select,
+textarea,
+input[type="text"],
+input[type="password"],
+input[type="datetime"],
+input[type="datetime-local"],
+input[type="date"],
+input[type="month"],
+input[type="time"],
+input[type="week"],
+input[type="number"],
+input[type="email"],
+input[type="url"],
+input[type="search"],
+input[type="tel"],
+input[type="color"],
+.uneditable-input {
+ display: inline-block;
+ height: 18px;
+ padding: 4px;
+ margin-bottom: 9px;
+ font-size: 12px;
+ line-height: 18px;
+ color: #555555;
+}
+input,
+textarea {
+ width: 210px;
+}
+textarea {
+ height: auto;
+}
+textarea,
+input[type="text"],
+input[type="password"],
+input[type="datetime"],
+input[type="datetime-local"],
+input[type="date"],
+input[type="month"],
+input[type="time"],
+input[type="week"],
+input[type="number"],
+input[type="email"],
+input[type="url"],
+input[type="search"],
+input[type="tel"],
+input[type="color"],
+.uneditable-input {
+ background-color: #ffffff;
+ border: 1px solid #cccccc;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+ -webkit-box-shadow: inset 0 1px 1px rgba(0, 0, 0, 0.075);
+ -moz-box-shadow: inset 0 1px 1px rgba(0, 0, 0, 0.075);
+ box-shadow: inset 0 1px 1px rgba(0, 0, 0, 0.075);
+ -webkit-transition: border linear 0.2s, box-shadow linear 0.2s;
+ -moz-transition: border linear 0.2s, box-shadow linear 0.2s;
+ -ms-transition: border linear 0.2s, box-shadow linear 0.2s;
+ -o-transition: border linear 0.2s, box-shadow linear 0.2s;
+ transition: border linear 0.2s, box-shadow linear 0.2s;
+}
+textarea:focus,
+input[type="text"]:focus,
+input[type="password"]:focus,
+input[type="datetime"]:focus,
+input[type="datetime-local"]:focus,
+input[type="date"]:focus,
+input[type="month"]:focus,
+input[type="time"]:focus,
+input[type="week"]:focus,
+input[type="number"]:focus,
+input[type="email"]:focus,
+input[type="url"]:focus,
+input[type="search"]:focus,
+input[type="tel"]:focus,
+input[type="color"]:focus,
+.uneditable-input:focus {
+ border-color: rgba(82, 168, 236, 0.8);
+ outline: 0;
+ outline: thin dotted \9;
+ /* IE6-9 */
+
+ -webkit-box-shadow: inset 0 1px 1px rgba(0,0,0,.075), 0 0 8px rgba(82,168,236,.6);
+ -moz-box-shadow: inset 0 1px 1px rgba(0,0,0,.075), 0 0 8px rgba(82,168,236,.6);
+ box-shadow: inset 0 1px 1px rgba(0,0,0,.075), 0 0 8px rgba(82,168,236,.6);
+}
+input[type="radio"],
+input[type="checkbox"] {
+ margin: 3px 0;
+ *margin-top: 0;
+ /* IE7 */
+
+ line-height: normal;
+ cursor: pointer;
+}
+input[type="submit"],
+input[type="reset"],
+input[type="button"],
+input[type="radio"],
+input[type="checkbox"] {
+ width: auto;
+}
+.uneditable-textarea {
+ width: auto;
+ height: auto;
+}
+select,
+input[type="file"] {
+ height: 28px;
+ /* In IE7, the height of the select element cannot be changed by height, only font-size */
+
+ *margin-top: 4px;
+ /* For IE7, add top margin to align select with labels */
+
+ line-height: 28px;
+}
+select {
+ width: 220px;
+ border: 1px solid #bbb;
+}
+select[multiple],
+select[size] {
+ height: auto;
+}
+select:focus,
+input[type="file"]:focus,
+input[type="radio"]:focus,
+input[type="checkbox"]:focus {
+ outline: thin dotted #333;
+ outline: 5px auto -webkit-focus-ring-color;
+ outline-offset: -2px;
+}
+.radio,
+.checkbox {
+ min-height: 18px;
+ padding-left: 18px;
+}
+.radio input[type="radio"],
+.checkbox input[type="checkbox"] {
+ float: left;
+ margin-left: -18px;
+}
+.controls > .radio:first-child,
+.controls > .checkbox:first-child {
+ padding-top: 5px;
+}
+.radio.inline,
+.checkbox.inline {
+ display: inline-block;
+ padding-top: 5px;
+ margin-bottom: 0;
+ vertical-align: middle;
+}
+.radio.inline + .radio.inline,
+.checkbox.inline + .checkbox.inline {
+ margin-left: 10px;
+}
+.input-mini {
+ width: 60px;
+}
+.input-small {
+ width: 90px;
+}
+.input-medium {
+ width: 150px;
+}
+.input-large {
+ width: 210px;
+}
+.input-xlarge {
+ width: 270px;
+}
+.input-xxlarge {
+ width: 530px;
+}
+input[class*="span"],
+select[class*="span"],
+textarea[class*="span"],
+.uneditable-input[class*="span"],
+.row-fluid input[class*="span"],
+.row-fluid select[class*="span"],
+.row-fluid textarea[class*="span"],
+.row-fluid .uneditable-input[class*="span"] {
+ float: none;
+ margin-left: 0;
+}
+.input-append input[class*="span"],
+.input-append .uneditable-input[class*="span"],
+.input-prepend input[class*="span"],
+.input-prepend .uneditable-input[class*="span"],
+.row-fluid .input-prepend [class*="span"],
+.row-fluid .input-append [class*="span"] {
+ display: inline-block;
+}
+input,
+textarea,
+.uneditable-input {
+ margin-left: 0;
+}
+input.span12, textarea.span12, .uneditable-input.span12 {
+ width: 930px;
+}
+input.span11, textarea.span11, .uneditable-input.span11 {
+ width: 850px;
+}
+input.span10, textarea.span10, .uneditable-input.span10 {
+ width: 770px;
+}
+input.span9, textarea.span9, .uneditable-input.span9 {
+ width: 690px;
+}
+input.span8, textarea.span8, .uneditable-input.span8 {
+ width: 610px;
+}
+input.span7, textarea.span7, .uneditable-input.span7 {
+ width: 530px;
+}
+input.span6, textarea.span6, .uneditable-input.span6 {
+ width: 450px;
+}
+input.span5, textarea.span5, .uneditable-input.span5 {
+ width: 370px;
+}
+input.span4, textarea.span4, .uneditable-input.span4 {
+ width: 290px;
+}
+input.span3, textarea.span3, .uneditable-input.span3 {
+ width: 210px;
+}
+input.span2, textarea.span2, .uneditable-input.span2 {
+ width: 130px;
+}
+input.span1, textarea.span1, .uneditable-input.span1 {
+ width: 50px;
+}
+input[disabled],
+select[disabled],
+textarea[disabled],
+input[readonly],
+select[readonly],
+textarea[readonly] {
+ cursor: not-allowed;
+ background-color: #eeeeee;
+ border-color: #ddd;
+}
+input[type="radio"][disabled],
+input[type="checkbox"][disabled],
+input[type="radio"][readonly],
+input[type="checkbox"][readonly] {
+ background-color: transparent;
+}
+.control-group.warning > label,
+.control-group.warning .help-block,
+.control-group.warning .help-inline {
+ color: #c09853;
+}
+.control-group.warning .checkbox,
+.control-group.warning .radio,
+.control-group.warning input,
+.control-group.warning select,
+.control-group.warning textarea {
+ color: #c09853;
+ border-color: #c09853;
+}
+.control-group.warning .checkbox:focus,
+.control-group.warning .radio:focus,
+.control-group.warning input:focus,
+.control-group.warning select:focus,
+.control-group.warning textarea:focus {
+ border-color: #a47e3c;
+ -webkit-box-shadow: 0 0 6px #dbc59e;
+ -moz-box-shadow: 0 0 6px #dbc59e;
+ box-shadow: 0 0 6px #dbc59e;
+}
+.control-group.warning .input-prepend .add-on,
+.control-group.warning .input-append .add-on {
+ color: #c09853;
+ background-color: #fcf8e3;
+ border-color: #c09853;
+}
+.control-group.error > label,
+.control-group.error .help-block,
+.control-group.error .help-inline {
+ color: #b94a48;
+}
+.control-group.error .checkbox,
+.control-group.error .radio,
+.control-group.error input,
+.control-group.error select,
+.control-group.error textarea {
+ color: #b94a48;
+ border-color: #b94a48;
+}
+.control-group.error .checkbox:focus,
+.control-group.error .radio:focus,
+.control-group.error input:focus,
+.control-group.error select:focus,
+.control-group.error textarea:focus {
+ border-color: #953b39;
+ -webkit-box-shadow: 0 0 6px #d59392;
+ -moz-box-shadow: 0 0 6px #d59392;
+ box-shadow: 0 0 6px #d59392;
+}
+.control-group.error .input-prepend .add-on,
+.control-group.error .input-append .add-on {
+ color: #b94a48;
+ background-color: #f2dede;
+ border-color: #b94a48;
+}
+.control-group.success > label,
+.control-group.success .help-block,
+.control-group.success .help-inline {
+ color: #468847;
+}
+.control-group.success .checkbox,
+.control-group.success .radio,
+.control-group.success input,
+.control-group.success select,
+.control-group.success textarea {
+ color: #468847;
+ border-color: #468847;
+}
+.control-group.success .checkbox:focus,
+.control-group.success .radio:focus,
+.control-group.success input:focus,
+.control-group.success select:focus,
+.control-group.success textarea:focus {
+ border-color: #356635;
+ -webkit-box-shadow: 0 0 6px #7aba7b;
+ -moz-box-shadow: 0 0 6px #7aba7b;
+ box-shadow: 0 0 6px #7aba7b;
+}
+.control-group.success .input-prepend .add-on,
+.control-group.success .input-append .add-on {
+ color: #468847;
+ background-color: #dff0d8;
+ border-color: #468847;
+}
+input:focus:required:invalid,
+textarea:focus:required:invalid,
+select:focus:required:invalid {
+ color: #b94a48;
+ border-color: #ee5f5b;
+}
+input:focus:required:invalid:focus,
+textarea:focus:required:invalid:focus,
+select:focus:required:invalid:focus {
+ border-color: #e9322d;
+ -webkit-box-shadow: 0 0 6px #f8b9b7;
+ -moz-box-shadow: 0 0 6px #f8b9b7;
+ box-shadow: 0 0 6px #f8b9b7;
+}
+.form-actions {
+ padding: 17px 20px 18px;
+ margin-top: 18px;
+ margin-bottom: 18px;
+ background-color: #f5f5f5;
+ border-top: 1px solid #e5e5e5;
+ *zoom: 1;
+}
+.form-actions:before,
+.form-actions:after {
+ display: table;
+ content: "";
+}
+.form-actions:after {
+ clear: both;
+}
+.uneditable-input {
+ overflow: hidden;
+ white-space: nowrap;
+ cursor: not-allowed;
+ background-color: #ffffff;
+ border-color: #eee;
+ -webkit-box-shadow: inset 0 1px 2px rgba(0, 0, 0, 0.025);
+ -moz-box-shadow: inset 0 1px 2px rgba(0, 0, 0, 0.025);
+ box-shadow: inset 0 1px 2px rgba(0, 0, 0, 0.025);
+}
+:-moz-placeholder {
+ color: #999999;
+}
+:-ms-input-placeholder {
+ color: #999999;
+}
+::-webkit-input-placeholder {
+ color: #999999;
+}
+.help-block,
+.help-inline {
+ color: #555555;
+}
+.help-block {
+ display: block;
+ margin-bottom: 9px;
+}
+.help-inline {
+ display: inline-block;
+ *display: inline;
+ /* IE7 inline-block hack */
+
+ *zoom: 1;
+ vertical-align: middle;
+ padding-left: 5px;
+}
+.input-prepend,
+.input-append {
+ margin-bottom: 5px;
+}
+.input-prepend input,
+.input-append input,
+.input-prepend select,
+.input-append select,
+.input-prepend .uneditable-input,
+.input-append .uneditable-input {
+ position: relative;
+ margin-bottom: 0;
+ *margin-left: 0;
+ vertical-align: middle;
+ -webkit-border-radius: 0 3px 3px 0;
+ -moz-border-radius: 0 3px 3px 0;
+ border-radius: 0 3px 3px 0;
+}
+.input-prepend input:focus,
+.input-append input:focus,
+.input-prepend select:focus,
+.input-append select:focus,
+.input-prepend .uneditable-input:focus,
+.input-append .uneditable-input:focus {
+ z-index: 2;
+}
+.input-prepend .uneditable-input,
+.input-append .uneditable-input {
+ border-left-color: #ccc;
+}
+.input-prepend .add-on,
+.input-append .add-on {
+ display: inline-block;
+ width: auto;
+ height: 18px;
+ min-width: 16px;
+ padding: 4px 5px;
+ font-weight: normal;
+ line-height: 18px;
+ text-align: center;
+ text-shadow: 0 1px 0 #ffffff;
+ vertical-align: middle;
+ background-color: #eeeeee;
+ border: 1px solid #ccc;
+}
+.input-prepend .add-on,
+.input-append .add-on,
+.input-prepend .btn,
+.input-append .btn {
+ margin-left: -1px;
+ -webkit-border-radius: 0;
+ -moz-border-radius: 0;
+ border-radius: 0;
+}
+.input-prepend .active,
+.input-append .active {
+ background-color: #a9dba9;
+ border-color: #46a546;
+}
+.input-prepend .add-on,
+.input-prepend .btn {
+ margin-right: -1px;
+}
+.input-prepend .add-on:first-child,
+.input-prepend .btn:first-child {
+ -webkit-border-radius: 3px 0 0 3px;
+ -moz-border-radius: 3px 0 0 3px;
+ border-radius: 3px 0 0 3px;
+}
+.input-append input,
+.input-append select,
+.input-append .uneditable-input {
+ -webkit-border-radius: 3px 0 0 3px;
+ -moz-border-radius: 3px 0 0 3px;
+ border-radius: 3px 0 0 3px;
+}
+.input-append .uneditable-input {
+ border-right-color: #ccc;
+ border-left-color: #eee;
+}
+.input-append .add-on:last-child,
+.input-append .btn:last-child {
+ -webkit-border-radius: 0 3px 3px 0;
+ -moz-border-radius: 0 3px 3px 0;
+ border-radius: 0 3px 3px 0;
+}
+.input-prepend.input-append input,
+.input-prepend.input-append select,
+.input-prepend.input-append .uneditable-input {
+ -webkit-border-radius: 0;
+ -moz-border-radius: 0;
+ border-radius: 0;
+}
+.input-prepend.input-append .add-on:first-child,
+.input-prepend.input-append .btn:first-child {
+ margin-right: -1px;
+ -webkit-border-radius: 3px 0 0 3px;
+ -moz-border-radius: 3px 0 0 3px;
+ border-radius: 3px 0 0 3px;
+}
+.input-prepend.input-append .add-on:last-child,
+.input-prepend.input-append .btn:last-child {
+ margin-left: -1px;
+ -webkit-border-radius: 0 3px 3px 0;
+ -moz-border-radius: 0 3px 3px 0;
+ border-radius: 0 3px 3px 0;
+}
+.search-query {
+ padding-right: 14px;
+ padding-right: 4px \9;
+ padding-left: 14px;
+ padding-left: 4px \9;
+ /* IE7-8 doesn't have border-radius, so don't indent the padding */
+
+ margin-bottom: 0;
+ -webkit-border-radius: 14px;
+ -moz-border-radius: 14px;
+ border-radius: 14px;
+}
+.form-search input,
+.form-inline input,
+.form-horizontal input,
+.form-search textarea,
+.form-inline textarea,
+.form-horizontal textarea,
+.form-search select,
+.form-inline select,
+.form-horizontal select,
+.form-search .help-inline,
+.form-inline .help-inline,
+.form-horizontal .help-inline,
+.form-search .uneditable-input,
+.form-inline .uneditable-input,
+.form-horizontal .uneditable-input,
+.form-search .input-prepend,
+.form-inline .input-prepend,
+.form-horizontal .input-prepend,
+.form-search .input-append,
+.form-inline .input-append,
+.form-horizontal .input-append {
+ display: inline-block;
+ *display: inline;
+ /* IE7 inline-block hack */
+
+ *zoom: 1;
+ margin-bottom: 0;
+}
+.form-search .hide,
+.form-inline .hide,
+.form-horizontal .hide {
+ display: none;
+}
+.form-search label,
+.form-inline label {
+ display: inline-block;
+}
+.form-search .input-append,
+.form-inline .input-append,
+.form-search .input-prepend,
+.form-inline .input-prepend {
+ margin-bottom: 0;
+}
+.form-search .radio,
+.form-search .checkbox,
+.form-inline .radio,
+.form-inline .checkbox {
+ padding-left: 0;
+ margin-bottom: 0;
+ vertical-align: middle;
+}
+.form-search .radio input[type="radio"],
+.form-search .checkbox input[type="checkbox"],
+.form-inline .radio input[type="radio"],
+.form-inline .checkbox input[type="checkbox"] {
+ float: left;
+ margin-right: 3px;
+ margin-left: 0;
+}
+.control-group {
+ margin-bottom: 9px;
+}
+legend + .control-group {
+ margin-top: 18px;
+ -webkit-margin-top-collapse: separate;
+}
+.form-horizontal .control-group {
+ margin-bottom: 18px;
+ *zoom: 1;
+}
+.form-horizontal .control-group:before,
+.form-horizontal .control-group:after {
+ display: table;
+ content: "";
+}
+.form-horizontal .control-group:after {
+ clear: both;
+}
+.form-horizontal .control-label {
+ float: left;
+ width: 140px;
+ padding-top: 5px;
+ text-align: right;
+}
+.form-horizontal .controls {
+ *display: inline-block;
+ *padding-left: 20px;
+ margin-left: 160px;
+ *margin-left: 0;
+}
+.form-horizontal .controls:first-child {
+ *padding-left: 160px;
+}
+.form-horizontal .help-block {
+ margin-top: 9px;
+ margin-bottom: 0;
+}
+.form-horizontal .form-actions {
+ padding-left: 160px;
+}
+.btn {
+ display: inline-block;
+ *display: inline;
+ /* IE7 inline-block hack */
+
+ *zoom: 1;
+ padding: 4px 10px 4px;
+ margin-bottom: 0;
+ font-size: 10px;
+ line-height: 18px;
+ *line-height: 20px;
+ color: #333333;
+ text-align: center;
+ text-shadow: 0 1px 1px rgba(255, 255, 255, 0.75);
+ vertical-align: middle;
+ cursor: pointer;
+ background-color: #f5f5f5;
+ background-image: -moz-linear-gradient(top, #ffffff, #e6e6e6);
+ background-image: -ms-linear-gradient(top, #ffffff, #e6e6e6);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#ffffff), to(#e6e6e6));
+ background-image: -webkit-linear-gradient(top, #ffffff, #e6e6e6);
+ background-image: -o-linear-gradient(top, #ffffff, #e6e6e6);
+ background-image: linear-gradient(top, #ffffff, #e6e6e6);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#ffffff', endColorstr='#e6e6e6', GradientType=0);
+ border-color: #e6e6e6 #e6e6e6 #bfbfbf;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #e6e6e6;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+ border: 1px solid #cccccc;
+ *border: 0;
+ border-bottom-color: #b3b3b3;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+ *margin-left: .3em;
+ -webkit-box-shadow: inset 0 1px 0 rgba(255,255,255,.2), 0 1px 2px rgba(0,0,0,.05);
+ -moz-box-shadow: inset 0 1px 0 rgba(255,255,255,.2), 0 1px 2px rgba(0,0,0,.05);
+ box-shadow: inset 0 1px 0 rgba(255,255,255,.2), 0 1px 2px rgba(0,0,0,.05);
+}
+.btn:hover,
+.btn:active,
+.btn.active,
+.btn.disabled,
+.btn[disabled] {
+ background-color: #e6e6e6;
+ *background-color: #d9d9d9;
+}
+.btn:active,
+.btn.active {
+ background-color: #cccccc \9;
+}
+.btn:first-child {
+ *margin-left: 0;
+}
+.btn:hover {
+ color: #333333;
+ text-decoration: none;
+ background-color: #e6e6e6;
+ *background-color: #d9d9d9;
+ /* Buttons in IE7 don't get borders, so darken on hover */
+
+ background-position: 0 -15px;
+ -webkit-transition: background-position 0.1s linear;
+ -moz-transition: background-position 0.1s linear;
+ -ms-transition: background-position 0.1s linear;
+ -o-transition: background-position 0.1s linear;
+ transition: background-position 0.1s linear;
+}
+.btn:focus {
+ outline: thin dotted #333;
+ outline: 5px auto -webkit-focus-ring-color;
+ outline-offset: -2px;
+}
+.btn.active,
+.btn:active {
+ background-color: #e6e6e6;
+ background-color: #d9d9d9 \9;
+ background-image: none;
+ outline: 0;
+ -webkit-box-shadow: inset 0 2px 4px rgba(0,0,0,.15), 0 1px 2px rgba(0,0,0,.05);
+ -moz-box-shadow: inset 0 2px 4px rgba(0,0,0,.15), 0 1px 2px rgba(0,0,0,.05);
+ box-shadow: inset 0 2px 4px rgba(0,0,0,.15), 0 1px 2px rgba(0,0,0,.05);
+}
+.btn.disabled,
+.btn[disabled] {
+ cursor: default;
+ background-color: #e6e6e6;
+ background-image: none;
+ opacity: 0.65;
+ filter: alpha(opacity=65);
+ -webkit-box-shadow: none;
+ -moz-box-shadow: none;
+ box-shadow: none;
+}
+.btn-large {
+ padding: 9px 14px;
+ font-size: 15px;
+ line-height: normal;
+ -webkit-border-radius: 5px;
+ -moz-border-radius: 5px;
+ border-radius: 5px;
+}
+.btn-large [class^="icon-"] {
+ margin-top: 1px;
+}
+.btn-small {
+ padding: 5px 9px;
+ font-size: 11px;
+ line-height: 16px;
+}
+.btn-small [class^="icon-"] {
+ margin-top: -1px;
+}
+.btn-mini {
+ padding: 2px 6px;
+ font-size: 11px;
+ line-height: 14px;
+}
+.btn-primary,
+.btn-primary:hover,
+.btn-warning,
+.btn-warning:hover,
+.btn-danger,
+.btn-danger:hover,
+.btn-success,
+.btn-success:hover,
+.btn-info,
+.btn-info:hover,
+.btn-inverse,
+.btn-inverse:hover {
+ color: #ffffff;
+ text-shadow: 0 -1px 0 rgba(0, 0, 0, 0.25);
+}
+.btn-primary.active,
+.btn-warning.active,
+.btn-danger.active,
+.btn-success.active,
+.btn-info.active,
+.btn-inverse.active {
+ color: rgba(255, 255, 255, 0.75);
+}
+.btn {
+ border-color: #ccc;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+}
+.btn-primary {
+ background-color: #0074cc;
+ background-image: -moz-linear-gradient(top, #0088cc, #0055cc);
+ background-image: -ms-linear-gradient(top, #0088cc, #0055cc);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#0088cc), to(#0055cc));
+ background-image: -webkit-linear-gradient(top, #0088cc, #0055cc);
+ background-image: -o-linear-gradient(top, #0088cc, #0055cc);
+ background-image: linear-gradient(top, #0088cc, #0055cc);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#0088cc', endColorstr='#0055cc', GradientType=0);
+ border-color: #0055cc #0055cc #003580;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #0055cc;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+}
+.btn-primary:hover,
+.btn-primary:active,
+.btn-primary.active,
+.btn-primary.disabled,
+.btn-primary[disabled] {
+ background-color: #0055cc;
+ *background-color: #004ab3;
+}
+.btn-primary:active,
+.btn-primary.active {
+ background-color: #004099 \9;
+}
+.btn-warning {
+ background-color: #faa732;
+ background-image: -moz-linear-gradient(top, #fbb450, #f89406);
+ background-image: -ms-linear-gradient(top, #fbb450, #f89406);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#fbb450), to(#f89406));
+ background-image: -webkit-linear-gradient(top, #fbb450, #f89406);
+ background-image: -o-linear-gradient(top, #fbb450, #f89406);
+ background-image: linear-gradient(top, #fbb450, #f89406);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#fbb450', endColorstr='#f89406', GradientType=0);
+ border-color: #f89406 #f89406 #ad6704;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #f89406;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+}
+.btn-warning:hover,
+.btn-warning:active,
+.btn-warning.active,
+.btn-warning.disabled,
+.btn-warning[disabled] {
+ background-color: #f89406;
+ *background-color: #df8505;
+}
+.btn-warning:active,
+.btn-warning.active {
+ background-color: #c67605 \9;
+}
+.btn-danger {
+ background-color: #da4f49;
+ background-image: -moz-linear-gradient(top, #ee5f5b, #bd362f);
+ background-image: -ms-linear-gradient(top, #ee5f5b, #bd362f);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#ee5f5b), to(#bd362f));
+ background-image: -webkit-linear-gradient(top, #ee5f5b, #bd362f);
+ background-image: -o-linear-gradient(top, #ee5f5b, #bd362f);
+ background-image: linear-gradient(top, #ee5f5b, #bd362f);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#ee5f5b', endColorstr='#bd362f', GradientType=0);
+ border-color: #bd362f #bd362f #802420;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #bd362f;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+}
+.btn-danger:hover,
+.btn-danger:active,
+.btn-danger.active,
+.btn-danger.disabled,
+.btn-danger[disabled] {
+ background-color: #bd362f;
+ *background-color: #a9302a;
+}
+.btn-danger:active,
+.btn-danger.active {
+ background-color: #942a25 \9;
+}
+.btn-success {
+ background-color: #5bb75b;
+ background-image: -moz-linear-gradient(top, #62c462, #51a351);
+ background-image: -ms-linear-gradient(top, #62c462, #51a351);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#62c462), to(#51a351));
+ background-image: -webkit-linear-gradient(top, #62c462, #51a351);
+ background-image: -o-linear-gradient(top, #62c462, #51a351);
+ background-image: linear-gradient(top, #62c462, #51a351);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#62c462', endColorstr='#51a351', GradientType=0);
+ border-color: #51a351 #51a351 #387038;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #51a351;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+}
+.btn-success:hover,
+.btn-success:active,
+.btn-success.active,
+.btn-success.disabled,
+.btn-success[disabled] {
+ background-color: #51a351;
+ *background-color: #499249;
+}
+.btn-success:active,
+.btn-success.active {
+ background-color: #408140 \9;
+}
+.btn-info {
+ background-color: #49afcd;
+ background-image: -moz-linear-gradient(top, #5bc0de, #2f96b4);
+ background-image: -ms-linear-gradient(top, #5bc0de, #2f96b4);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#5bc0de), to(#2f96b4));
+ background-image: -webkit-linear-gradient(top, #5bc0de, #2f96b4);
+ background-image: -o-linear-gradient(top, #5bc0de, #2f96b4);
+ background-image: linear-gradient(top, #5bc0de, #2f96b4);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#5bc0de', endColorstr='#2f96b4', GradientType=0);
+ border-color: #2f96b4 #2f96b4 #1f6377;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #2f96b4;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+}
+.btn-info:hover,
+.btn-info:active,
+.btn-info.active,
+.btn-info.disabled,
+.btn-info[disabled] {
+ background-color: #2f96b4;
+ *background-color: #2a85a0;
+}
+.btn-info:active,
+.btn-info.active {
+ background-color: #24748c \9;
+}
+.btn-inverse {
+ background-color: #414141;
+ background-image: -moz-linear-gradient(top, #555555, #222222);
+ background-image: -ms-linear-gradient(top, #555555, #222222);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#555555), to(#222222));
+ background-image: -webkit-linear-gradient(top, #555555, #222222);
+ background-image: -o-linear-gradient(top, #555555, #222222);
+ background-image: linear-gradient(top, #555555, #222222);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#555555', endColorstr='#222222', GradientType=0);
+ border-color: #222222 #222222 #000000;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #222222;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+}
+.btn-inverse:hover,
+.btn-inverse:active,
+.btn-inverse.active,
+.btn-inverse.disabled,
+.btn-inverse[disabled] {
+ background-color: #222222;
+ *background-color: #151515;
+}
+.btn-inverse:active,
+.btn-inverse.active {
+ background-color: #080808 \9;
+}
+button.btn,
+input[type="submit"].btn {
+ *padding-top: 2px;
+ *padding-bottom: 2px;
+}
+button.btn::-moz-focus-inner,
+input[type="submit"].btn::-moz-focus-inner {
+ padding: 0;
+ border: 0;
+}
+button.btn.btn-large,
+input[type="submit"].btn.btn-large {
+ *padding-top: 7px;
+ *padding-bottom: 7px;
+}
+button.btn.btn-small,
+input[type="submit"].btn.btn-small {
+ *padding-top: 3px;
+ *padding-bottom: 3px;
+}
+button.btn.btn-mini,
+input[type="submit"].btn.btn-mini {
+ *padding-top: 1px;
+ *padding-bottom: 1px;
+}
+.btn-group {
+ position: relative;
+ *zoom: 1;
+ *margin-left: .3em;
+}
+.btn-group:before,
+.btn-group:after {
+ display: table;
+ content: "";
+}
+.btn-group:after {
+ clear: both;
+}
+.btn-group:first-child {
+ *margin-left: 0;
+}
+.btn-group + .btn-group {
+ margin-left: 5px;
+}
+.btn-toolbar {
+ margin-top: 9px;
+ margin-bottom: 9px;
+}
+.btn-toolbar .btn-group {
+ display: inline-block;
+ *display: inline;
+ /* IE7 inline-block hack */
+
+ *zoom: 1;
+}
+.btn-group > .btn {
+ position: relative;
+ float: left;
+ margin-left: -1px;
+ -webkit-border-radius: 0;
+ -moz-border-radius: 0;
+ border-radius: 0;
+}
+.btn-group > .btn:first-child {
+ margin-left: 0;
+ -webkit-border-top-left-radius: 4px;
+ -moz-border-radius-topleft: 4px;
+ border-top-left-radius: 4px;
+ -webkit-border-bottom-left-radius: 4px;
+ -moz-border-radius-bottomleft: 4px;
+ border-bottom-left-radius: 4px;
+}
+.btn-group > .btn:last-child,
+.btn-group > .dropdown-toggle {
+ -webkit-border-top-right-radius: 4px;
+ -moz-border-radius-topright: 4px;
+ border-top-right-radius: 4px;
+ -webkit-border-bottom-right-radius: 4px;
+ -moz-border-radius-bottomright: 4px;
+ border-bottom-right-radius: 4px;
+}
+.btn-group > .btn.large:first-child {
+ margin-left: 0;
+ -webkit-border-top-left-radius: 6px;
+ -moz-border-radius-topleft: 6px;
+ border-top-left-radius: 6px;
+ -webkit-border-bottom-left-radius: 6px;
+ -moz-border-radius-bottomleft: 6px;
+ border-bottom-left-radius: 6px;
+}
+.btn-group > .btn.large:last-child,
+.btn-group > .large.dropdown-toggle {
+ -webkit-border-top-right-radius: 6px;
+ -moz-border-radius-topright: 6px;
+ border-top-right-radius: 6px;
+ -webkit-border-bottom-right-radius: 6px;
+ -moz-border-radius-bottomright: 6px;
+ border-bottom-right-radius: 6px;
+}
+.btn-group > .btn:hover,
+.btn-group > .btn:focus,
+.btn-group > .btn:active,
+.btn-group > .btn.active {
+ z-index: 2;
+}
+.btn-group .dropdown-toggle:active,
+.btn-group.open .dropdown-toggle {
+ outline: 0;
+}
+.btn-group > .dropdown-toggle {
+ padding-left: 8px;
+ padding-right: 8px;
+ -webkit-box-shadow: inset 1px 0 0 rgba(255,255,255,.125), inset 0 1px 0 rgba(255,255,255,.2), 0 1px 2px rgba(0,0,0,.05);
+ -moz-box-shadow: inset 1px 0 0 rgba(255,255,255,.125), inset 0 1px 0 rgba(255,255,255,.2), 0 1px 2px rgba(0,0,0,.05);
+ box-shadow: inset 1px 0 0 rgba(255,255,255,.125), inset 0 1px 0 rgba(255,255,255,.2), 0 1px 2px rgba(0,0,0,.05);
+ *padding-top: 4px;
+ *padding-bottom: 4px;
+}
+.btn-group > .btn-mini.dropdown-toggle {
+ padding-left: 5px;
+ padding-right: 5px;
+}
+.btn-group > .btn-small.dropdown-toggle {
+ *padding-top: 4px;
+ *padding-bottom: 4px;
+}
+.btn-group > .btn-large.dropdown-toggle {
+ padding-left: 12px;
+ padding-right: 12px;
+}
+.btn-group.open .dropdown-toggle {
+ background-image: none;
+ -webkit-box-shadow: inset 0 2px 4px rgba(0,0,0,.15), 0 1px 2px rgba(0,0,0,.05);
+ -moz-box-shadow: inset 0 2px 4px rgba(0,0,0,.15), 0 1px 2px rgba(0,0,0,.05);
+ box-shadow: inset 0 2px 4px rgba(0,0,0,.15), 0 1px 2px rgba(0,0,0,.05);
+}
+.btn-group.open .btn.dropdown-toggle {
+ background-color: #e6e6e6;
+}
+.btn-group.open .btn-primary.dropdown-toggle {
+ background-color: #0055cc;
+}
+.btn-group.open .btn-warning.dropdown-toggle {
+ background-color: #f89406;
+}
+.btn-group.open .btn-danger.dropdown-toggle {
+ background-color: #bd362f;
+}
+.btn-group.open .btn-success.dropdown-toggle {
+ background-color: #51a351;
+}
+.btn-group.open .btn-info.dropdown-toggle {
+ background-color: #2f96b4;
+}
+.btn-group.open .btn-inverse.dropdown-toggle {
+ background-color: #222222;
+}
+.btn .caret {
+ margin-top: 7px;
+ margin-left: 0;
+}
+.btn:hover .caret,
+.open.btn-group .caret {
+ opacity: 1;
+ filter: alpha(opacity=100);
+}
+.btn-mini .caret {
+ margin-top: 5px;
+}
+.btn-small .caret {
+ margin-top: 6px;
+}
+.btn-large .caret {
+ margin-top: 6px;
+ border-left-width: 5px;
+ border-right-width: 5px;
+ border-top-width: 5px;
+}
+.dropup .btn-large .caret {
+ border-bottom: 5px solid #000000;
+ border-top: 0;
+}
+.btn-primary .caret,
+.btn-warning .caret,
+.btn-danger .caret,
+.btn-info .caret,
+.btn-success .caret,
+.btn-inverse .caret {
+ border-top-color: #ffffff;
+ border-bottom-color: #ffffff;
+ opacity: 0.75;
+ filter: alpha(opacity=75);
+}
+.nav {
+ margin-left: 0;
+ margin-bottom: 18px;
+ list-style: none;
+}
+.nav > li > a {
+ display: block;
+}
+.nav > li > a:hover {
+ text-decoration: none;
+ background-color: #eeeeee;
+}
+.nav > .pull-right {
+ float: right;
+}
+.nav .nav-header {
+ display: block;
+ padding: 3px 15px;
+ font-size: 11px;
+ font-weight: bold;
+ line-height: 18px;
+ color: #999999;
+ text-shadow: 0 1px 0 rgba(255, 255, 255, 0.5);
+ text-transform: uppercase;
+}
+.nav li + .nav-header {
+ margin-top: 9px;
+}
+.nav-list {
+ padding-left: 15px;
+ padding-right: 15px;
+ margin-bottom: 0;
+}
+.nav-list > li > a,
+.nav-list .nav-header {
+ margin-left: -15px;
+ margin-right: -15px;
+ text-shadow: 0 1px 0 rgba(255, 255, 255, 0.5);
+}
+.nav-list > li > a {
+ padding: 3px 15px;
+}
+.nav-list > .active > a,
+.nav-list > .active > a:hover {
+ color: #ffffff;
+ text-shadow: 0 -1px 0 rgba(0, 0, 0, 0.2);
+ background-color: #0088cc;
+}
+.nav-list [class^="icon-"] {
+ margin-right: 2px;
+}
+.nav-list .divider {
+ *width: 100%;
+ height: 1px;
+ margin: 8px 1px;
+ *margin: -5px 0 5px;
+ overflow: hidden;
+ background-color: #e5e5e5;
+ border-bottom: 1px solid #ffffff;
+}
+.nav-tabs,
+.nav-pills {
+ *zoom: 1;
+}
+.nav-tabs:before,
+.nav-pills:before,
+.nav-tabs:after,
+.nav-pills:after {
+ display: table;
+ content: "";
+}
+.nav-tabs:after,
+.nav-pills:after {
+ clear: both;
+}
+.nav-tabs > li,
+.nav-pills > li {
+ float: left;
+}
+.nav-tabs > li > a,
+.nav-pills > li > a {
+ padding-right: 12px;
+ padding-left: 12px;
+ margin-right: 2px;
+ line-height: 14px;
+}
+.nav-tabs {
+ border-bottom: 1px solid #ddd;
+}
+.nav-tabs > li {
+ margin-bottom: -1px;
+}
+.nav-tabs > li > a {
+ padding-top: 8px;
+ padding-bottom: 8px;
+ line-height: 18px;
+ border: 1px solid transparent;
+ -webkit-border-radius: 4px 4px 0 0;
+ -moz-border-radius: 4px 4px 0 0;
+ border-radius: 4px 4px 0 0;
+}
+.nav-tabs > li > a:hover {
+ border-color: #eeeeee #eeeeee #dddddd;
+}
+.nav-tabs > .active > a,
+.nav-tabs > .active > a:hover {
+ color: #555555;
+ background-color: #ffffff;
+ border: 1px solid #ddd;
+ border-bottom-color: transparent;
+ cursor: default;
+}
+.nav-pills > li > a {
+ padding-top: 8px;
+ padding-bottom: 8px;
+ margin-top: 2px;
+ margin-bottom: 2px;
+ -webkit-border-radius: 5px;
+ -moz-border-radius: 5px;
+ border-radius: 5px;
+}
+.nav-pills > .active > a,
+.nav-pills > .active > a:hover {
+ color: #ffffff;
+ background-color: #0088cc;
+}
+.nav-stacked > li {
+ float: none;
+}
+.nav-stacked > li > a {
+ margin-right: 0;
+}
+.nav-tabs.nav-stacked {
+ border-bottom: 0;
+}
+.nav-tabs.nav-stacked > li > a {
+ border: 1px solid #ddd;
+ -webkit-border-radius: 0;
+ -moz-border-radius: 0;
+ border-radius: 0;
+}
+.nav-tabs.nav-stacked > li:first-child > a {
+ -webkit-border-radius: 4px 4px 0 0;
+ -moz-border-radius: 4px 4px 0 0;
+ border-radius: 4px 4px 0 0;
+}
+.nav-tabs.nav-stacked > li:last-child > a {
+ -webkit-border-radius: 0 0 4px 4px;
+ -moz-border-radius: 0 0 4px 4px;
+ border-radius: 0 0 4px 4px;
+}
+.nav-tabs.nav-stacked > li > a:hover {
+ border-color: #ddd;
+ z-index: 2;
+}
+.nav-pills.nav-stacked > li > a {
+ margin-bottom: 3px;
+}
+.nav-pills.nav-stacked > li:last-child > a {
+ margin-bottom: 1px;
+}
+.nav-tabs .dropdown-menu {
+ -webkit-border-radius: 0 0 5px 5px;
+ -moz-border-radius: 0 0 5px 5px;
+ border-radius: 0 0 5px 5px;
+}
+.nav-pills .dropdown-menu {
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+.nav-tabs .dropdown-toggle .caret,
+.nav-pills .dropdown-toggle .caret {
+ border-top-color: #0088cc;
+ border-bottom-color: #0088cc;
+ margin-top: 6px;
+}
+.nav-tabs .dropdown-toggle:hover .caret,
+.nav-pills .dropdown-toggle:hover .caret {
+ border-top-color: #005580;
+ border-bottom-color: #005580;
+}
+.nav-tabs .active .dropdown-toggle .caret,
+.nav-pills .active .dropdown-toggle .caret {
+ border-top-color: #333333;
+ border-bottom-color: #333333;
+}
+.nav > .dropdown.active > a:hover {
+ color: #000000;
+ cursor: pointer;
+}
+.nav-tabs .open .dropdown-toggle,
+.nav-pills .open .dropdown-toggle,
+.nav > li.dropdown.open.active > a:hover {
+ color: #ffffff;
+ background-color: #999999;
+ border-color: #999999;
+}
+.nav li.dropdown.open .caret,
+.nav li.dropdown.open.active .caret,
+.nav li.dropdown.open a:hover .caret {
+ border-top-color: #ffffff;
+ border-bottom-color: #ffffff;
+ opacity: 1;
+ filter: alpha(opacity=100);
+}
+.tabs-stacked .open > a:hover {
+ border-color: #999999;
+}
+.tabbable {
+ *zoom: 1;
+}
+.tabbable:before,
+.tabbable:after {
+ display: table;
+ content: "";
+}
+.tabbable:after {
+ clear: both;
+}
+.tab-content {
+ overflow: auto;
+}
+.tabs-below > .nav-tabs,
+.tabs-right > .nav-tabs,
+.tabs-left > .nav-tabs {
+ border-bottom: 0;
+}
+.tab-content > .tab-pane,
+.pill-content > .pill-pane {
+ display: none;
+}
+.tab-content > .active,
+.pill-content > .active {
+ display: block;
+}
+.tabs-below > .nav-tabs {
+ border-top: 1px solid #ddd;
+}
+.tabs-below > .nav-tabs > li {
+ margin-top: -1px;
+ margin-bottom: 0;
+}
+.tabs-below > .nav-tabs > li > a {
+ -webkit-border-radius: 0 0 4px 4px;
+ -moz-border-radius: 0 0 4px 4px;
+ border-radius: 0 0 4px 4px;
+}
+.tabs-below > .nav-tabs > li > a:hover {
+ border-bottom-color: transparent;
+ border-top-color: #ddd;
+}
+.tabs-below > .nav-tabs > .active > a,
+.tabs-below > .nav-tabs > .active > a:hover {
+ border-color: transparent #ddd #ddd #ddd;
+}
+.tabs-left > .nav-tabs > li,
+.tabs-right > .nav-tabs > li {
+ float: none;
+}
+.tabs-left > .nav-tabs > li > a,
+.tabs-right > .nav-tabs > li > a {
+ min-width: 74px;
+ margin-right: 0;
+ margin-bottom: 3px;
+}
+.tabs-left > .nav-tabs {
+ float: left;
+ margin-right: 19px;
+ border-right: 1px solid #ddd;
+}
+.tabs-left > .nav-tabs > li > a {
+ margin-right: -1px;
+ -webkit-border-radius: 4px 0 0 4px;
+ -moz-border-radius: 4px 0 0 4px;
+ border-radius: 4px 0 0 4px;
+}
+.tabs-left > .nav-tabs > li > a:hover {
+ border-color: #eeeeee #dddddd #eeeeee #eeeeee;
+}
+.tabs-left > .nav-tabs .active > a,
+.tabs-left > .nav-tabs .active > a:hover {
+ border-color: #ddd transparent #ddd #ddd;
+ *border-right-color: #ffffff;
+}
+.tabs-right > .nav-tabs {
+ float: right;
+ margin-left: 19px;
+ border-left: 1px solid #ddd;
+}
+.tabs-right > .nav-tabs > li > a {
+ margin-left: -1px;
+ -webkit-border-radius: 0 4px 4px 0;
+ -moz-border-radius: 0 4px 4px 0;
+ border-radius: 0 4px 4px 0;
+}
+.tabs-right > .nav-tabs > li > a:hover {
+ border-color: #eeeeee #eeeeee #eeeeee #dddddd;
+}
+.tabs-right > .nav-tabs .active > a,
+.tabs-right > .nav-tabs .active > a:hover {
+ border-color: #ddd #ddd #ddd transparent;
+ *border-left-color: #ffffff;
+}
+.navbar {
+ *position: relative;
+ *z-index: 2;
+ overflow: visible;
+ margin-bottom: 18px;
+}
+.navbar-inner {
+ min-height: 40px;
+ padding-left: 20px;
+ padding-right: 20px;
+ background-color: #465158;
+ background-image: -moz-linear-gradient(top, #333333, #222222);
+ background-image: -ms-linear-gradient(top, #333333, #222222);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#333333), to(#222222));
+ background-image: -webkit-linear-gradient(top, #333333, #222222);
+ background-image: -o-linear-gradient(top, #333333, #222222);
+ background-image: linear-gradient(top, #333333, #222222);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#333333', endColorstr='#222222', GradientType=0);
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+ -webkit-box-shadow: 0 1px 3px rgba(0,0,0,.25), inset 0 -1px 0 rgba(0,0,0,.1);
+ -moz-box-shadow: 0 1px 3px rgba(0,0,0,.25), inset 0 -1px 0 rgba(0,0,0,.1);
+ box-shadow: 0 1px 3px rgba(0,0,0,.25), inset 0 -1px 0 rgba(0,0,0,.1);
+}
+.navbar .container {
+ width: auto;
+}
+.nav-collapse.collapse {
+ height: auto;
+}
+.navbar {
+ color: #999999;
+}
+.navbar .brand:hover {
+ text-decoration: none;
+}
+.navbar .brand {
+ float: left;
+ display: block;
+ padding: 8px 20px 12px;
+ margin-left: -20px;
+ font-size: 20px;
+ font-weight: 200;
+ line-height: 1;
+ color: #999999;
+}
+.navbar .navbar-text {
+ margin-bottom: 0;
+ line-height: 40px;
+}
+.navbar .navbar-link {
+ color: #999999;
+}
+.navbar .navbar-link:hover {
+ color: #ffffff;
+}
+.navbar .btn,
+.navbar .btn-group {
+ margin-top: 5px;
+}
+.navbar .btn-group .btn {
+ margin: 0;
+}
+.navbar-form {
+ margin-bottom: 0;
+ *zoom: 1;
+}
+.navbar-form:before,
+.navbar-form:after {
+ display: table;
+ content: "";
+}
+.navbar-form:after {
+ clear: both;
+}
+.navbar-form input,
+.navbar-form select,
+.navbar-form .radio,
+.navbar-form .checkbox {
+ margin-top: 5px;
+}
+.navbar-form input,
+.navbar-form select {
+ display: inline-block;
+ margin-bottom: 0;
+}
+.navbar-form input[type="image"],
+.navbar-form input[type="checkbox"],
+.navbar-form input[type="radio"] {
+ margin-top: 3px;
+}
+.navbar-form .input-append,
+.navbar-form .input-prepend {
+ margin-top: 6px;
+ white-space: nowrap;
+}
+.navbar-form .input-append input,
+.navbar-form .input-prepend input {
+ margin-top: 0;
+}
+.navbar-search {
+ position: relative;
+ float: left;
+ margin-top: 6px;
+ margin-bottom: 0;
+}
+.navbar-search .search-query {
+ padding: 4px 9px;
+ font-family: "Helvetica Neue", Helvetica, Arial, sans-serif;
+ font-size: 13px;
+ font-weight: normal;
+ line-height: 1;
+ color: #ffffff;
+ background-color: #626262;
+ border: 1px solid #151515;
+ -webkit-box-shadow: inset 0 1px 2px rgba(0,0,0,.1), 0 1px 0 rgba(255,255,255,.15);
+ -moz-box-shadow: inset 0 1px 2px rgba(0,0,0,.1), 0 1px 0 rgba(255,255,255,.15);
+ box-shadow: inset 0 1px 2px rgba(0,0,0,.1), 0 1px 0 rgba(255,255,255,.15);
+ -webkit-transition: none;
+ -moz-transition: none;
+ -ms-transition: none;
+ -o-transition: none;
+ transition: none;
+}
+.navbar-search .search-query:-moz-placeholder {
+ color: #cccccc;
+}
+.navbar-search .search-query:-ms-input-placeholder {
+ color: #cccccc;
+}
+.navbar-search .search-query::-webkit-input-placeholder {
+ color: #cccccc;
+}
+.navbar-search .search-query:focus,
+.navbar-search .search-query.focused {
+ padding: 5px 10px;
+ color: #333333;
+ text-shadow: 0 1px 0 #ffffff;
+ background-color: #ffffff;
+ border: 0;
+ -webkit-box-shadow: 0 0 3px rgba(0, 0, 0, 0.15);
+ -moz-box-shadow: 0 0 3px rgba(0, 0, 0, 0.15);
+ box-shadow: 0 0 3px rgba(0, 0, 0, 0.15);
+ outline: 0;
+}
+.navbar-fixed-top,
+.navbar-fixed-bottom {
+ position: fixed;
+ right: 0;
+ left: 0;
+ z-index: 1030;
+ margin-bottom: 0;
+}
+.navbar-fixed-top .navbar-inner,
+.navbar-fixed-bottom .navbar-inner {
+ padding-left: 0;
+ padding-right: 0;
+ -webkit-border-radius: 0;
+ -moz-border-radius: 0;
+ border-radius: 0;
+}
+.navbar-fixed-top .container,
+.navbar-fixed-bottom .container {
+ width: 940px;
+}
+.navbar-fixed-top {
+ top: 0;
+}
+.navbar-fixed-bottom {
+ bottom: 0;
+}
+.navbar .nav {
+ position: relative;
+ left: 0;
+ display: block;
+ float: left;
+ margin: 0 10px 0 0;
+}
+.navbar .nav.pull-right {
+ float: right;
+}
+.navbar .nav > li {
+ display: block;
+ float: left;
+}
+.navbar .nav > li > a {
+ float: none;
+ padding: 9px 10px 11px;
+ line-height: 19px;
+ color: #999999;
+ text-decoration: none;
+ text-shadow: 0 -1px 0 rgba(0, 0, 0, 0.25);
+}
+.navbar .btn {
+ display: inline-block;
+ padding: 4px 10px 4px;
+ margin: 5px 5px 6px;
+ line-height: 18px;
+}
+.navbar .btn-group {
+ margin: 0;
+ padding: 5px 5px 6px;
+}
+.navbar .nav > li > a:hover {
+ background-color: transparent;
+ color: #ffffff;
+ text-decoration: none;
+}
+.navbar .nav .active > a,
+.navbar .nav .active > a:hover {
+ color: #ffffff;
+ text-decoration: none;
+ background-color: #222222;
+}
+.navbar .divider-vertical {
+ height: 40px;
+ width: 1px;
+ margin: 0 9px;
+ overflow: hidden;
+ background-color: #222222;
+ border-right: 1px solid #333333;
+}
+.navbar .nav.pull-right {
+ margin-left: 10px;
+ margin-right: 0;
+}
+.navbar .btn-navbar {
+ display: none;
+ float: right;
+ padding: 7px 10px;
+ margin-left: 5px;
+ margin-right: 5px;
+ background-color: #2c2c2c;
+ background-image: -moz-linear-gradient(top, #333333, #222222);
+ background-image: -ms-linear-gradient(top, #333333, #222222);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#333333), to(#222222));
+ background-image: -webkit-linear-gradient(top, #333333, #222222);
+ background-image: -o-linear-gradient(top, #333333, #222222);
+ background-image: linear-gradient(top, #333333, #222222);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#333333', endColorstr='#222222', GradientType=0);
+ border-color: #222222 #222222 #000000;
+ border-color: rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.1) rgba(0, 0, 0, 0.25);
+ *background-color: #222222;
+ /* Darken IE7 buttons by default so they stand out more given they won't have borders */
+
+ filter: progid:DXImageTransform.Microsoft.gradient(enabled = false);
+ -webkit-box-shadow: inset 0 1px 0 rgba(255,255,255,.1), 0 1px 0 rgba(255,255,255,.075);
+ -moz-box-shadow: inset 0 1px 0 rgba(255,255,255,.1), 0 1px 0 rgba(255,255,255,.075);
+ box-shadow: inset 0 1px 0 rgba(255,255,255,.1), 0 1px 0 rgba(255,255,255,.075);
+}
+.navbar .btn-navbar:hover,
+.navbar .btn-navbar:active,
+.navbar .btn-navbar.active,
+.navbar .btn-navbar.disabled,
+.navbar .btn-navbar[disabled] {
+ background-color: #222222;
+ *background-color: #151515;
+}
+.navbar .btn-navbar:active,
+.navbar .btn-navbar.active {
+ background-color: #080808 \9;
+}
+.navbar .btn-navbar .icon-bar {
+ display: block;
+ width: 18px;
+ height: 2px;
+ background-color: #f5f5f5;
+ -webkit-border-radius: 1px;
+ -moz-border-radius: 1px;
+ border-radius: 1px;
+ -webkit-box-shadow: 0 1px 0 rgba(0, 0, 0, 0.25);
+ -moz-box-shadow: 0 1px 0 rgba(0, 0, 0, 0.25);
+ box-shadow: 0 1px 0 rgba(0, 0, 0, 0.25);
+}
+.btn-navbar .icon-bar + .icon-bar {
+ margin-top: 3px;
+}
+.navbar .dropdown-menu:before {
+ content: '';
+ display: inline-block;
+ border-left: 7px solid transparent;
+ border-right: 7px solid transparent;
+ border-bottom: 7px solid #ccc;
+ border-bottom-color: rgba(0, 0, 0, 0.2);
+ position: absolute;
+ top: -7px;
+ left: 9px;
+}
+.navbar .dropdown-menu:after {
+ content: '';
+ display: inline-block;
+ border-left: 6px solid transparent;
+ border-right: 6px solid transparent;
+ border-bottom: 6px solid #ffffff;
+ position: absolute;
+ top: -6px;
+ left: 10px;
+}
+.navbar-fixed-bottom .dropdown-menu:before {
+ border-top: 7px solid #ccc;
+ border-top-color: rgba(0, 0, 0, 0.2);
+ border-bottom: 0;
+ bottom: -7px;
+ top: auto;
+}
+.navbar-fixed-bottom .dropdown-menu:after {
+ border-top: 6px solid #ffffff;
+ border-bottom: 0;
+ bottom: -6px;
+ top: auto;
+}
+.navbar .nav li.dropdown .dropdown-toggle .caret,
+.navbar .nav li.dropdown.open .caret {
+ border-top-color: #ffffff;
+ border-bottom-color: #ffffff;
+}
+.navbar .nav li.dropdown.active .caret {
+ opacity: 1;
+ filter: alpha(opacity=100);
+}
+.navbar .nav li.dropdown.open > .dropdown-toggle,
+.navbar .nav li.dropdown.active > .dropdown-toggle,
+.navbar .nav li.dropdown.open.active > .dropdown-toggle {
+ background-color: transparent;
+}
+.navbar .nav li.dropdown.active > .dropdown-toggle:hover {
+ color: #ffffff;
+}
+.navbar .pull-right .dropdown-menu,
+.navbar .dropdown-menu.pull-right {
+ left: auto;
+ right: 0;
+}
+.navbar .pull-right .dropdown-menu:before,
+.navbar .dropdown-menu.pull-right:before {
+ left: auto;
+ right: 12px;
+}
+.navbar .pull-right .dropdown-menu:after,
+.navbar .dropdown-menu.pull-right:after {
+ left: auto;
+ right: 13px;
+}
+.breadcrumb {
+ padding: 7px 14px;
+ margin: 0 0 18px;
+ list-style: none;
+ background-color: #fbfbfb;
+ background-image: -moz-linear-gradient(top, #ffffff, #f5f5f5);
+ background-image: -ms-linear-gradient(top, #ffffff, #f5f5f5);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#ffffff), to(#f5f5f5));
+ background-image: -webkit-linear-gradient(top, #ffffff, #f5f5f5);
+ background-image: -o-linear-gradient(top, #ffffff, #f5f5f5);
+ background-image: linear-gradient(top, #ffffff, #f5f5f5);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#ffffff', endColorstr='#f5f5f5', GradientType=0);
+ border: 1px solid #ddd;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+ -webkit-box-shadow: inset 0 1px 0 #ffffff;
+ -moz-box-shadow: inset 0 1px 0 #ffffff;
+ box-shadow: inset 0 1px 0 #ffffff;
+}
+.breadcrumb li {
+ display: inline-block;
+ *display: inline;
+ /* IE7 inline-block hack */
+
+ *zoom: 1;
+ text-shadow: 0 1px 0 #ffffff;
+}
+.breadcrumb .divider {
+ padding: 0 5px;
+ color: #999999;
+}
+.breadcrumb .active a {
+ color: #333333;
+}
+.pagination {
+ height: 36px;
+ margin: 18px 0;
+}
+.pagination ul {
+ display: inline-block;
+ *display: inline;
+ /* IE7 inline-block hack */
+
+ *zoom: 1;
+ margin-left: 0;
+ margin-bottom: 0;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+ -webkit-box-shadow: 0 1px 2px rgba(0, 0, 0, 0.05);
+ -moz-box-shadow: 0 1px 2px rgba(0, 0, 0, 0.05);
+ box-shadow: 0 1px 2px rgba(0, 0, 0, 0.05);
+}
+.pagination li {
+ display: inline;
+}
+.pagination a {
+ float: left;
+ padding: 0 14px;
+ line-height: 34px;
+ text-decoration: none;
+ border: 1px solid #ddd;
+ border-left-width: 0;
+}
+.pagination a:hover,
+.pagination .active a {
+ background-color: #f5f5f5;
+}
+.pagination .active a {
+ color: #999999;
+ cursor: default;
+}
+.pagination .disabled span,
+.pagination .disabled a,
+.pagination .disabled a:hover {
+ color: #999999;
+ background-color: transparent;
+ cursor: default;
+}
+.pagination li:first-child a {
+ border-left-width: 1px;
+ -webkit-border-radius: 3px 0 0 3px;
+ -moz-border-radius: 3px 0 0 3px;
+ border-radius: 3px 0 0 3px;
+}
+.pagination li:last-child a {
+ -webkit-border-radius: 0 3px 3px 0;
+ -moz-border-radius: 0 3px 3px 0;
+ border-radius: 0 3px 3px 0;
+}
+.pagination-centered {
+ text-align: center;
+}
+.pagination-right {
+ text-align: right;
+}
+.pager {
+ margin-left: 0;
+ margin-bottom: 18px;
+ list-style: none;
+ text-align: center;
+ *zoom: 1;
+}
+.pager:before,
+.pager:after {
+ display: table;
+ content: "";
+}
+.pager:after {
+ clear: both;
+}
+.pager li {
+ display: inline;
+}
+.pager a {
+ display: inline-block;
+ padding: 5px 14px;
+ background-color: #fff;
+ border: 1px solid #ddd;
+ -webkit-border-radius: 15px;
+ -moz-border-radius: 15px;
+ border-radius: 15px;
+}
+.pager a:hover {
+ text-decoration: none;
+ background-color: #f5f5f5;
+}
+.pager .next a {
+ float: right;
+}
+.pager .previous a {
+ float: left;
+}
+.pager .disabled a,
+.pager .disabled a:hover {
+ color: #999999;
+ background-color: #fff;
+ cursor: default;
+}
+.thumbnails {
+ margin-left: -20px;
+ list-style: none;
+ *zoom: 1;
+}
+.thumbnails:before,
+.thumbnails:after {
+ display: table;
+ content: "";
+}
+.thumbnails:after {
+ clear: both;
+}
+.row-fluid .thumbnails {
+ margin-left: 0;
+}
+.thumbnails > li {
+ float: left;
+ margin-bottom: 18px;
+ margin-left: 20px;
+}
+.thumbnail {
+ display: block;
+ padding: 4px;
+ line-height: 1;
+ border: 1px solid #ddd;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+ -webkit-box-shadow: 0 1px 1px rgba(0, 0, 0, 0.075);
+ -moz-box-shadow: 0 1px 1px rgba(0, 0, 0, 0.075);
+ box-shadow: 0 1px 1px rgba(0, 0, 0, 0.075);
+}
+a.thumbnail:hover {
+ border-color: #0088cc;
+ -webkit-box-shadow: 0 1px 4px rgba(0, 105, 214, 0.25);
+ -moz-box-shadow: 0 1px 4px rgba(0, 105, 214, 0.25);
+ box-shadow: 0 1px 4px rgba(0, 105, 214, 0.25);
+}
+.thumbnail > img {
+ display: block;
+ max-width: 100%;
+ margin-left: auto;
+ margin-right: auto;
+}
+.thumbnail .caption {
+ padding: 9px;
+}
+.alert {
+ padding: 8px 35px 8px 14px;
+ margin-bottom: 18px;
+ text-shadow: 0 1px 0 rgba(255, 255, 255, 0.5);
+ background-color: #fcf8e3;
+ border: 1px solid #fbeed5;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+ color: #c09853;
+}
+.alert-heading {
+ color: inherit;
+}
+.alert .close {
+ position: relative;
+ top: -2px;
+ right: -21px;
+ line-height: 18px;
+}
+.alert-success {
+ background-color: #dff0d8;
+ border-color: #d6e9c6;
+ color: #468847;
+}
+.alert-danger,
+.alert-error {
+ background-color: #f2dede;
+ border-color: #eed3d7;
+ color: #b94a48;
+}
+.alert-info {
+ background-color: #d9edf7;
+ border-color: #bce8f1;
+ color: #3a87ad;
+}
+.alert-block {
+ padding-top: 14px;
+ padding-bottom: 14px;
+}
+.alert-block > p,
+.alert-block > ul {
+ margin-bottom: 0;
+}
+.alert-block p + p {
+ margin-top: 5px;
+}
+@-webkit-keyframes progress-bar-stripes {
+ from {
+ background-position: 40px 0;
+ }
+ to {
+ background-position: 0 0;
+ }
+}
+@-moz-keyframes progress-bar-stripes {
+ from {
+ background-position: 40px 0;
+ }
+ to {
+ background-position: 0 0;
+ }
+}
+@-ms-keyframes progress-bar-stripes {
+ from {
+ background-position: 40px 0;
+ }
+ to {
+ background-position: 0 0;
+ }
+}
+@-o-keyframes progress-bar-stripes {
+ from {
+ background-position: 0 0;
+ }
+ to {
+ background-position: 40px 0;
+ }
+}
+@keyframes progress-bar-stripes {
+ from {
+ background-position: 40px 0;
+ }
+ to {
+ background-position: 0 0;
+ }
+}
+.progress {
+ overflow: hidden;
+ height: 18px;
+ margin-bottom: 18px;
+ background-color: #f7f7f7;
+ background-image: -moz-linear-gradient(top, #f5f5f5, #f9f9f9);
+ background-image: -ms-linear-gradient(top, #f5f5f5, #f9f9f9);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#f5f5f5), to(#f9f9f9));
+ background-image: -webkit-linear-gradient(top, #f5f5f5, #f9f9f9);
+ background-image: -o-linear-gradient(top, #f5f5f5, #f9f9f9);
+ background-image: linear-gradient(top, #f5f5f5, #f9f9f9);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#f5f5f5', endColorstr='#f9f9f9', GradientType=0);
+ -webkit-box-shadow: inset 0 1px 2px rgba(0, 0, 0, 0.1);
+ -moz-box-shadow: inset 0 1px 2px rgba(0, 0, 0, 0.1);
+ box-shadow: inset 0 1px 2px rgba(0, 0, 0, 0.1);
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+.progress .bar {
+ width: 0%;
+ height: 18px;
+ color: #ffffff;
+ font-size: 12px;
+ text-align: center;
+ text-shadow: 0 -1px 0 rgba(0, 0, 0, 0.25);
+ background-color: #0e90d2;
+ background-image: -moz-linear-gradient(top, #149bdf, #0480be);
+ background-image: -ms-linear-gradient(top, #149bdf, #0480be);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#149bdf), to(#0480be));
+ background-image: -webkit-linear-gradient(top, #149bdf, #0480be);
+ background-image: -o-linear-gradient(top, #149bdf, #0480be);
+ background-image: linear-gradient(top, #149bdf, #0480be);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#149bdf', endColorstr='#0480be', GradientType=0);
+ -webkit-box-shadow: inset 0 -1px 0 rgba(0, 0, 0, 0.15);
+ -moz-box-shadow: inset 0 -1px 0 rgba(0, 0, 0, 0.15);
+ box-shadow: inset 0 -1px 0 rgba(0, 0, 0, 0.15);
+ -webkit-box-sizing: border-box;
+ -moz-box-sizing: border-box;
+ -ms-box-sizing: border-box;
+ box-sizing: border-box;
+ -webkit-transition: width 0.6s ease;
+ -moz-transition: width 0.6s ease;
+ -ms-transition: width 0.6s ease;
+ -o-transition: width 0.6s ease;
+ transition: width 0.6s ease;
+}
+.progress-striped .bar {
+ background-color: #149bdf;
+ background-image: -webkit-gradient(linear, 0 100%, 100% 0, color-stop(0.25, rgba(255, 255, 255, 0.15)), color-stop(0.25, transparent), color-stop(0.5, transparent), color-stop(0.5, rgba(255, 255, 255, 0.15)), color-stop(0.75, rgba(255, 255, 255, 0.15)), color-stop(0.75, transparent), to(transparent));
+ background-image: -webkit-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -moz-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -ms-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -o-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ -webkit-background-size: 40px 40px;
+ -moz-background-size: 40px 40px;
+ -o-background-size: 40px 40px;
+ background-size: 40px 40px;
+}
+.progress.active .bar {
+ -webkit-animation: progress-bar-stripes 2s linear infinite;
+ -moz-animation: progress-bar-stripes 2s linear infinite;
+ -ms-animation: progress-bar-stripes 2s linear infinite;
+ -o-animation: progress-bar-stripes 2s linear infinite;
+ animation: progress-bar-stripes 2s linear infinite;
+}
+.progress-danger .bar {
+ background-color: #dd514c;
+ background-image: -moz-linear-gradient(top, #ee5f5b, #c43c35);
+ background-image: -ms-linear-gradient(top, #ee5f5b, #c43c35);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#ee5f5b), to(#c43c35));
+ background-image: -webkit-linear-gradient(top, #ee5f5b, #c43c35);
+ background-image: -o-linear-gradient(top, #ee5f5b, #c43c35);
+ background-image: linear-gradient(top, #ee5f5b, #c43c35);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#ee5f5b', endColorstr='#c43c35', GradientType=0);
+}
+.progress-danger.progress-striped .bar {
+ background-color: #ee5f5b;
+ background-image: -webkit-gradient(linear, 0 100%, 100% 0, color-stop(0.25, rgba(255, 255, 255, 0.15)), color-stop(0.25, transparent), color-stop(0.5, transparent), color-stop(0.5, rgba(255, 255, 255, 0.15)), color-stop(0.75, rgba(255, 255, 255, 0.15)), color-stop(0.75, transparent), to(transparent));
+ background-image: -webkit-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -moz-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -ms-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -o-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+}
+.progress-success .bar {
+ background-color: #5eb95e;
+ background-image: -moz-linear-gradient(top, #62c462, #57a957);
+ background-image: -ms-linear-gradient(top, #62c462, #57a957);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#62c462), to(#57a957));
+ background-image: -webkit-linear-gradient(top, #62c462, #57a957);
+ background-image: -o-linear-gradient(top, #62c462, #57a957);
+ background-image: linear-gradient(top, #62c462, #57a957);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#62c462', endColorstr='#57a957', GradientType=0);
+}
+.progress-success.progress-striped .bar {
+ background-color: #62c462;
+ background-image: -webkit-gradient(linear, 0 100%, 100% 0, color-stop(0.25, rgba(255, 255, 255, 0.15)), color-stop(0.25, transparent), color-stop(0.5, transparent), color-stop(0.5, rgba(255, 255, 255, 0.15)), color-stop(0.75, rgba(255, 255, 255, 0.15)), color-stop(0.75, transparent), to(transparent));
+ background-image: -webkit-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -moz-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -ms-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -o-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+}
+.progress-info .bar {
+ background-color: #4bb1cf;
+ background-image: -moz-linear-gradient(top, #5bc0de, #339bb9);
+ background-image: -ms-linear-gradient(top, #5bc0de, #339bb9);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#5bc0de), to(#339bb9));
+ background-image: -webkit-linear-gradient(top, #5bc0de, #339bb9);
+ background-image: -o-linear-gradient(top, #5bc0de, #339bb9);
+ background-image: linear-gradient(top, #5bc0de, #339bb9);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#5bc0de', endColorstr='#339bb9', GradientType=0);
+}
+.progress-info.progress-striped .bar {
+ background-color: #5bc0de;
+ background-image: -webkit-gradient(linear, 0 100%, 100% 0, color-stop(0.25, rgba(255, 255, 255, 0.15)), color-stop(0.25, transparent), color-stop(0.5, transparent), color-stop(0.5, rgba(255, 255, 255, 0.15)), color-stop(0.75, rgba(255, 255, 255, 0.15)), color-stop(0.75, transparent), to(transparent));
+ background-image: -webkit-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -moz-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -ms-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -o-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+}
+.progress-warning .bar {
+ background-color: #faa732;
+ background-image: -moz-linear-gradient(top, #fbb450, #f89406);
+ background-image: -ms-linear-gradient(top, #fbb450, #f89406);
+ background-image: -webkit-gradient(linear, 0 0, 0 100%, from(#fbb450), to(#f89406));
+ background-image: -webkit-linear-gradient(top, #fbb450, #f89406);
+ background-image: -o-linear-gradient(top, #fbb450, #f89406);
+ background-image: linear-gradient(top, #fbb450, #f89406);
+ background-repeat: repeat-x;
+ filter: progid:DXImageTransform.Microsoft.gradient(startColorstr='#fbb450', endColorstr='#f89406', GradientType=0);
+}
+.progress-warning.progress-striped .bar {
+ background-color: #fbb450;
+ background-image: -webkit-gradient(linear, 0 100%, 100% 0, color-stop(0.25, rgba(255, 255, 255, 0.15)), color-stop(0.25, transparent), color-stop(0.5, transparent), color-stop(0.5, rgba(255, 255, 255, 0.15)), color-stop(0.75, rgba(255, 255, 255, 0.15)), color-stop(0.75, transparent), to(transparent));
+ background-image: -webkit-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -moz-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -ms-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: -o-linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+ background-image: linear-gradient(-45deg, rgba(255, 255, 255, 0.15) 25%, transparent 25%, transparent 50%, rgba(255, 255, 255, 0.15) 50%, rgba(255, 255, 255, 0.15) 75%, transparent 75%, transparent);
+}
+.hero-unit {
+ padding: 60px;
+ margin-bottom: 30px;
+ background-color: #eeeeee;
+ -webkit-border-radius: 6px;
+ -moz-border-radius: 6px;
+ border-radius: 6px;
+}
+.hero-unit h1 {
+ margin-bottom: 0;
+ font-size: 60px;
+ line-height: 1;
+ color: inherit;
+ letter-spacing: -1px;
+}
+.hero-unit p {
+ font-size: 18px;
+ font-weight: 200;
+ line-height: 27px;
+ color: inherit;
+}
+.tooltip {
+ position: absolute;
+ z-index: 1020;
+ display: block;
+ visibility: visible;
+ padding: 5px;
+ font-size: 11px;
+ opacity: 0;
+ filter: alpha(opacity=0);
+}
+.tooltip.in {
+ opacity: 0.8;
+ filter: alpha(opacity=80);
+}
+.tooltip.top {
+ margin-top: -2px;
+}
+.tooltip.right {
+ margin-left: 2px;
+}
+.tooltip.bottom {
+ margin-top: 2px;
+}
+.tooltip.left {
+ margin-left: -2px;
+}
+.tooltip.top .tooltip-arrow {
+ bottom: 0;
+ left: 50%;
+ margin-left: -5px;
+ border-left: 5px solid transparent;
+ border-right: 5px solid transparent;
+ border-top: 5px solid #000000;
+}
+.tooltip.left .tooltip-arrow {
+ top: 50%;
+ right: 0;
+ margin-top: -5px;
+ border-top: 5px solid transparent;
+ border-bottom: 5px solid transparent;
+ border-left: 5px solid #000000;
+}
+.tooltip.bottom .tooltip-arrow {
+ top: 0;
+ left: 50%;
+ margin-left: -5px;
+ border-left: 5px solid transparent;
+ border-right: 5px solid transparent;
+ border-bottom: 5px solid #000000;
+}
+.tooltip.right .tooltip-arrow {
+ top: 50%;
+ left: 0;
+ margin-top: -5px;
+ border-top: 5px solid transparent;
+ border-bottom: 5px solid transparent;
+ border-right: 5px solid #000000;
+}
+.tooltip-inner {
+ max-width: 200px;
+ padding: 3px 8px;
+ color: #ffffff;
+ text-align: center;
+ text-decoration: none;
+ background-color: #000000;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+.tooltip-arrow {
+ position: absolute;
+ width: 0;
+ height: 0;
+}
+.popover {
+ position: absolute;
+ top: 0;
+ left: 0;
+ z-index: 1010;
+ display: none;
+ padding: 5px;
+}
+.popover.top {
+ margin-top: -5px;
+}
+.popover.right {
+ margin-left: 5px;
+}
+.popover.bottom {
+ margin-top: 5px;
+}
+.popover.left {
+ margin-left: -5px;
+}
+.popover.top .arrow {
+ bottom: 0;
+ left: 50%;
+ margin-left: -5px;
+ border-left: 5px solid transparent;
+ border-right: 5px solid transparent;
+ border-top: 5px solid #000000;
+}
+.popover.right .arrow {
+ top: 50%;
+ left: 0;
+ margin-top: -5px;
+ border-top: 5px solid transparent;
+ border-bottom: 5px solid transparent;
+ border-right: 5px solid #000000;
+}
+.popover.bottom .arrow {
+ top: 0;
+ left: 50%;
+ margin-left: -5px;
+ border-left: 5px solid transparent;
+ border-right: 5px solid transparent;
+ border-bottom: 5px solid #000000;
+}
+.popover.left .arrow {
+ top: 50%;
+ right: 0;
+ margin-top: -5px;
+ border-top: 5px solid transparent;
+ border-bottom: 5px solid transparent;
+ border-left: 5px solid #000000;
+}
+.popover .arrow {
+ position: absolute;
+ width: 0;
+ height: 0;
+}
+.popover-inner {
+ padding: 3px;
+ width: 280px;
+ overflow: hidden;
+ background: #000000;
+ background: rgba(0, 0, 0, 0.8);
+ -webkit-border-radius: 6px;
+ -moz-border-radius: 6px;
+ border-radius: 6px;
+ -webkit-box-shadow: 0 3px 7px rgba(0, 0, 0, 0.3);
+ -moz-box-shadow: 0 3px 7px rgba(0, 0, 0, 0.3);
+ box-shadow: 0 3px 7px rgba(0, 0, 0, 0.3);
+}
+.popover-title {
+ padding: 9px 15px;
+ line-height: 1;
+ background-color: #f5f5f5;
+ border-bottom: 1px solid #eee;
+ -webkit-border-radius: 3px 3px 0 0;
+ -moz-border-radius: 3px 3px 0 0;
+ border-radius: 3px 3px 0 0;
+}
+.popover-content {
+ padding: 14px;
+ background-color: #ffffff;
+ -webkit-border-radius: 0 0 3px 3px;
+ -moz-border-radius: 0 0 3px 3px;
+ border-radius: 0 0 3px 3px;
+ -webkit-background-clip: padding-box;
+ -moz-background-clip: padding-box;
+ background-clip: padding-box;
+}
+.popover-content p,
+.popover-content ul,
+.popover-content ol {
+ margin-bottom: 0;
+}
+.modal-open .dropdown-menu {
+ z-index: 2050;
+}
+.modal-open .dropdown.open {
+ *z-index: 2050;
+}
+.modal-open .popover {
+ z-index: 2060;
+}
+.modal-open .tooltip {
+ z-index: 2070;
+}
+.modal-backdrop {
+ position: fixed;
+ top: 0;
+ right: 0;
+ bottom: 0;
+ left: 0;
+ z-index: 1040;
+ background-color: #000000;
+}
+.modal-backdrop.fade {
+ opacity: 0;
+}
+.modal-backdrop,
+.modal-backdrop.fade.in {
+ opacity: 0.8;
+ filter: alpha(opacity=80);
+}
+.modal {
+ position: fixed;
+ top: 50%;
+ left: 50%;
+ z-index: 1050;
+ overflow: auto;
+ width: 560px;
+ margin: -250px 0 0 -280px;
+ background-color: #ffffff;
+ border: 1px solid #999;
+ border: 1px solid rgba(0, 0, 0, 0.3);
+ *border: 1px solid #999;
+ /* IE6-7 */
+
+ -webkit-border-radius: 6px;
+ -moz-border-radius: 6px;
+ border-radius: 6px;
+ -webkit-box-shadow: 0 3px 7px rgba(0, 0, 0, 0.3);
+ -moz-box-shadow: 0 3px 7px rgba(0, 0, 0, 0.3);
+ box-shadow: 0 3px 7px rgba(0, 0, 0, 0.3);
+ -webkit-background-clip: padding-box;
+ -moz-background-clip: padding-box;
+ background-clip: padding-box;
+}
+.modal.fade {
+ -webkit-transition: opacity .3s linear, top .3s ease-out;
+ -moz-transition: opacity .3s linear, top .3s ease-out;
+ -ms-transition: opacity .3s linear, top .3s ease-out;
+ -o-transition: opacity .3s linear, top .3s ease-out;
+ transition: opacity .3s linear, top .3s ease-out;
+ top: -25%;
+}
+.modal.fade.in {
+ top: 50%;
+}
+.modal-header {
+ padding: 9px 15px;
+ border-bottom: 1px solid #eee;
+}
+.modal-header .close {
+ margin-top: 2px;
+}
+.modal-body {
+ overflow-y: auto;
+ max-height: 400px;
+ padding: 15px;
+}
+.modal-form {
+ margin-bottom: 0;
+}
+.modal-footer {
+ padding: 14px 15px 15px;
+ margin-bottom: 0;
+ text-align: right;
+ background-color: #f5f5f5;
+ border-top: 1px solid #ddd;
+ -webkit-border-radius: 0 0 6px 6px;
+ -moz-border-radius: 0 0 6px 6px;
+ border-radius: 0 0 6px 6px;
+ -webkit-box-shadow: inset 0 1px 0 #ffffff;
+ -moz-box-shadow: inset 0 1px 0 #ffffff;
+ box-shadow: inset 0 1px 0 #ffffff;
+ *zoom: 1;
+}
+.modal-footer:before,
+.modal-footer:after {
+ display: table;
+ content: "";
+}
+.modal-footer:after {
+ clear: both;
+}
+.modal-footer .btn + .btn {
+ margin-left: 5px;
+ margin-bottom: 0;
+}
+.modal-footer .btn-group .btn + .btn {
+ margin-left: -1px;
+}
+.dropup,
+.dropdown {
+ position: relative;
+}
+.dropdown-toggle {
+ *margin-bottom: -3px;
+}
+.dropdown-toggle:active,
+.open .dropdown-toggle {
+ outline: 0;
+}
+.caret {
+ display: inline-block;
+ width: 0;
+ height: 0;
+ vertical-align: top;
+ border-top: 4px solid #000000;
+ border-right: 4px solid transparent;
+ border-left: 4px solid transparent;
+ content: "";
+ opacity: 0.3;
+ filter: alpha(opacity=30);
+}
+.dropdown .caret {
+ margin-top: 8px;
+ margin-left: 2px;
+}
+.dropdown:hover .caret,
+.open .caret {
+ opacity: 1;
+ filter: alpha(opacity=100);
+}
+.dropdown-menu {
+ position: absolute;
+ top: 100%;
+ left: 0;
+ z-index: 1000;
+ display: none;
+ float: left;
+ min-width: 160px;
+ padding: 4px 0;
+ margin: 1px 0 0;
+ list-style: none;
+ background-color: #ffffff;
+ border: 1px solid #ccc;
+ border: 1px solid rgba(0, 0, 0, 0.2);
+ *border-right-width: 2px;
+ *border-bottom-width: 2px;
+ -webkit-border-radius: 5px;
+ -moz-border-radius: 5px;
+ border-radius: 5px;
+ -webkit-box-shadow: 0 5px 10px rgba(0, 0, 0, 0.2);
+ -moz-box-shadow: 0 5px 10px rgba(0, 0, 0, 0.2);
+ box-shadow: 0 5px 10px rgba(0, 0, 0, 0.2);
+ -webkit-background-clip: padding-box;
+ -moz-background-clip: padding;
+ background-clip: padding-box;
+}
+.dropdown-menu.pull-right {
+ right: 0;
+ left: auto;
+}
+.dropdown-menu .divider {
+ *width: 100%;
+ height: 1px;
+ margin: 8px 1px;
+ *margin: -5px 0 5px;
+ overflow: hidden;
+ background-color: #e5e5e5;
+ border-bottom: 1px solid #ffffff;
+}
+.dropdown-menu a {
+ display: block;
+ padding: 3px 15px;
+ clear: both;
+ font-weight: normal;
+ line-height: 18px;
+ color: #333333;
+ white-space: nowrap;
+}
+.dropdown-menu li > a:hover,
+.dropdown-menu .active > a,
+.dropdown-menu .active > a:hover {
+ color: #ffffff;
+ text-decoration: none;
+ background-color: #0088cc;
+}
+.open {
+ *z-index: 1000;
+}
+.open > .dropdown-menu {
+ display: block;
+}
+.pull-right > .dropdown-menu {
+ right: 0;
+ left: auto;
+}
+.dropup .caret,
+.navbar-fixed-bottom .dropdown .caret {
+ border-top: 0;
+ border-bottom: 4px solid #000000;
+ content: "\2191";
+}
+.dropup .dropdown-menu,
+.navbar-fixed-bottom .dropdown .dropdown-menu {
+ top: auto;
+ bottom: 100%;
+ margin-bottom: 1px;
+}
+.typeahead {
+ margin-top: 2px;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+.accordion {
+ margin-bottom: 18px;
+}
+.accordion-group {
+ margin-bottom: 2px;
+ border: 1px solid #e5e5e5;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+}
+.accordion-heading {
+ border-bottom: 0;
+}
+.accordion-heading .accordion-toggle {
+ display: block;
+ padding: 8px 15px;
+}
+.accordion-toggle {
+ cursor: pointer;
+}
+.accordion-inner {
+ padding: 9px 15px;
+ border-top: 1px solid #e5e5e5;
+}
+.carousel {
+ position: relative;
+ margin-bottom: 18px;
+ line-height: 1;
+}
+.carousel-inner {
+ overflow: hidden;
+ width: 100%;
+ position: relative;
+}
+.carousel .item {
+ display: none;
+ position: relative;
+ -webkit-transition: 0.6s ease-in-out left;
+ -moz-transition: 0.6s ease-in-out left;
+ -ms-transition: 0.6s ease-in-out left;
+ -o-transition: 0.6s ease-in-out left;
+ transition: 0.6s ease-in-out left;
+}
+.carousel .item > img {
+ display: block;
+ line-height: 1;
+}
+.carousel .active,
+.carousel .next,
+.carousel .prev {
+ display: block;
+}
+.carousel .active {
+ left: 0;
+}
+.carousel .next,
+.carousel .prev {
+ position: absolute;
+ top: 0;
+ width: 100%;
+}
+.carousel .next {
+ left: 100%;
+}
+.carousel .prev {
+ left: -100%;
+}
+.carousel .next.left,
+.carousel .prev.right {
+ left: 0;
+}
+.carousel .active.left {
+ left: -100%;
+}
+.carousel .active.right {
+ left: 100%;
+}
+.carousel-control {
+ position: absolute;
+ top: 40%;
+ left: 15px;
+ width: 40px;
+ height: 40px;
+ margin-top: -20px;
+ font-size: 60px;
+ font-weight: 100;
+ line-height: 30px;
+ color: #ffffff;
+ text-align: center;
+ background: #222222;
+ border: 3px solid #ffffff;
+ -webkit-border-radius: 23px;
+ -moz-border-radius: 23px;
+ border-radius: 23px;
+ opacity: 0.5;
+ filter: alpha(opacity=50);
+}
+.carousel-control.right {
+ left: auto;
+ right: 15px;
+}
+.carousel-control:hover {
+ color: #ffffff;
+ text-decoration: none;
+ opacity: 0.9;
+ filter: alpha(opacity=90);
+}
+.carousel-caption {
+ position: absolute;
+ left: 0;
+ right: 0;
+ bottom: 0;
+ padding: 10px 15px 5px;
+ background: #333333;
+/* background: rgba(0, 0, 0, 0.75);*/
+}
+.carousel-caption h4,
+.carousel-caption p {
+ color: #ffffff;
+}
+.well {
+ min-height: 20px;
+ padding: 19px;
+ margin-bottom: 20px;
+ background-color: #f5f5f5;
+ border: 1px solid #eee;
+ border: 1px solid rgba(0, 0, 0, 0.05);
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+ -webkit-box-shadow: inset 0 1px 1px rgba(0, 0, 0, 0.05);
+ -moz-box-shadow: inset 0 1px 1px rgba(0, 0, 0, 0.05);
+ box-shadow: inset 0 1px 1px rgba(0, 0, 0, 0.05);
+}
+.well blockquote {
+ border-color: #ddd;
+ border-color: rgba(0, 0, 0, 0.15);
+}
+.well-large {
+ padding: 24px;
+ -webkit-border-radius: 6px;
+ -moz-border-radius: 6px;
+ border-radius: 6px;
+}
+.well-small {
+ padding: 9px;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+}
+.close {
+ float: right;
+ font-size: 20px;
+ font-weight: bold;
+ line-height: 18px;
+ color: #000000;
+ text-shadow: 0 1px 0 #ffffff;
+ opacity: 0.2;
+ filter: alpha(opacity=20);
+}
+.close:hover {
+ color: #000000;
+ text-decoration: none;
+ cursor: pointer;
+ opacity: 0.4;
+ filter: alpha(opacity=40);
+}
+button.close {
+ padding: 0;
+ cursor: pointer;
+ background: transparent;
+ border: 0;
+ -webkit-appearance: none;
+}
+.pull-right {
+ float: right;
+}
+.pull-left {
+ float: left;
+}
+.hide {
+ display: none;
+}
+.show {
+ display: block;
+}
+.invisible {
+ visibility: hidden;
+}
+.fade {
+ opacity: 0;
+ -webkit-transition: opacity 0.15s linear;
+ -moz-transition: opacity 0.15s linear;
+ -ms-transition: opacity 0.15s linear;
+ -o-transition: opacity 0.15s linear;
+ transition: opacity 0.15s linear;
+}
+.fade.in {
+ opacity: 1;
+}
+.collapse {
+ position: relative;
+ height: 0;
+ overflow: hidden;
+ -webkit-transition: height 0.35s ease;
+ -moz-transition: height 0.35s ease;
+ -ms-transition: height 0.35s ease;
+ -o-transition: height 0.35s ease;
+ transition: height 0.35s ease;
+}
+.collapse.in {
+ height: auto;
+}
+.hidden {
+ display: none;
+ visibility: hidden;
+}
+.visible-phone {
+ display: none !important;
+}
+.visible-tablet {
+ display: none !important;
+}
+.hidden-desktop {
+ display: none !important;
+}
+@media (max-width: 767px) {
+ .visible-phone {
+ display: inherit !important;
+ }
+ .hidden-phone {
+ display: none !important;
+ }
+ .hidden-desktop {
+ display: inherit !important;
+ }
+ .visible-desktop {
+ display: none !important;
+ }
+}
+@media (min-width: 768px) and (max-width: 979px) {
+ .visible-tablet {
+ display: inherit !important;
+ }
+ .hidden-tablet {
+ display: none !important;
+ }
+ .hidden-desktop {
+ display: inherit !important;
+ }
+ .visible-desktop {
+ display: none !important ;
+ }
+}
+@media (max-width: 480px) {
+ .nav-collapse {
+ -webkit-transform: translate3d(0, 0, 0);
+ }
+ .page-header h1 small {
+ display: block;
+ line-height: 18px;
+ }
+ input[type="checkbox"],
+ input[type="radio"] {
+ border: 1px solid #ccc;
+ }
+ .form-horizontal .control-group > label {
+ float: none;
+ width: auto;
+ padding-top: 0;
+ text-align: left;
+ }
+ .form-horizontal .controls {
+ margin-left: 0;
+ }
+ .form-horizontal .control-list {
+ padding-top: 0;
+ }
+ .form-horizontal .form-actions {
+ padding-left: 10px;
+ padding-right: 10px;
+ }
+ .modal {
+ position: absolute;
+ top: 10px;
+ left: 10px;
+ right: 10px;
+ width: auto;
+ margin: 0;
+ }
+ .modal.fade.in {
+ top: auto;
+ }
+ .modal-header .close {
+ padding: 10px;
+ margin: -10px;
+ }
+ .carousel-caption {
+ position: static;
+ }
+}
+@media (max-width: 767px) {
+ body {
+ padding-left: 20px;
+ padding-right: 20px;
+ }
+ .navbar-fixed-top,
+ .navbar-fixed-bottom {
+ margin-left: -20px;
+ margin-right: -20px;
+ }
+ .container-fluid {
+ padding: 0;
+ }
+ .dl-horizontal dt {
+ float: none;
+ clear: none;
+ width: auto;
+ text-align: left;
+ }
+ .dl-horizontal dd {
+ margin-left: 0;
+ }
+ .container {
+ width: auto;
+ }
+ .row-fluid {
+ width: 100%;
+ }
+ .row,
+ .thumbnails {
+ margin-left: 0;
+ }
+ [class*="span"],
+ .row-fluid [class*="span"] {
+ float: none;
+ display: block;
+ width: auto;
+ margin-left: 0;
+ }
+ .input-large,
+ .input-xlarge,
+ .input-xxlarge,
+ input[class*="span"],
+ select[class*="span"],
+ textarea[class*="span"],
+ .uneditable-input {
+ display: block;
+ width: 100%;
+ min-height: 28px;
+ -webkit-box-sizing: border-box;
+ -moz-box-sizing: border-box;
+ -ms-box-sizing: border-box;
+ box-sizing: border-box;
+ }
+ .input-prepend input,
+ .input-append input,
+ .input-prepend input[class*="span"],
+ .input-append input[class*="span"] {
+ display: inline-block;
+ width: auto;
+ }
+}
+@media (min-width: 768px) and (max-width: 979px) {
+ .row {
+ margin-left: -20px;
+ *zoom: 1;
+ }
+ .row:before,
+ .row:after {
+ display: table;
+ content: "";
+ }
+ .row:after {
+ clear: both;
+ }
+ [class*="span"] {
+ float: left;
+ margin-left: 20px;
+ }
+ .container,
+ .navbar-fixed-top .container,
+ .navbar-fixed-bottom .container {
+ width: 724px;
+ }
+ .span12 {
+ width: 724px;
+ }
+ .span11 {
+ width: 662px;
+ }
+ .span10 {
+ width: 600px;
+ }
+ .span9 {
+ width: 538px;
+ }
+ .span8 {
+ width: 476px;
+ }
+ .span7 {
+ width: 414px;
+ }
+ .span6 {
+ width: 352px;
+ }
+ .span5 {
+ width: 290px;
+ }
+ .span4 {
+ width: 228px;
+ }
+ .span3 {
+ width: 166px;
+ }
+ .span2 {
+ width: 104px;
+ }
+ .span1 {
+ width: 42px;
+ }
+ .offset12 {
+ margin-left: 764px;
+ }
+ .offset11 {
+ margin-left: 702px;
+ }
+ .offset10 {
+ margin-left: 640px;
+ }
+ .offset9 {
+ margin-left: 578px;
+ }
+ .offset8 {
+ margin-left: 516px;
+ }
+ .offset7 {
+ margin-left: 454px;
+ }
+ .offset6 {
+ margin-left: 392px;
+ }
+ .offset5 {
+ margin-left: 330px;
+ }
+ .offset4 {
+ margin-left: 268px;
+ }
+ .offset3 {
+ margin-left: 206px;
+ }
+ .offset2 {
+ margin-left: 144px;
+ }
+ .offset1 {
+ margin-left: 82px;
+ }
+ .row-fluid {
+ width: 100%;
+ *zoom: 1;
+ }
+ .row-fluid:before,
+ .row-fluid:after {
+ display: table;
+ content: "";
+ }
+ .row-fluid:after {
+ clear: both;
+ }
+ .row-fluid [class*="span"] {
+ display: block;
+ width: 100%;
+ min-height: 28px;
+ -webkit-box-sizing: border-box;
+ -moz-box-sizing: border-box;
+ -ms-box-sizing: border-box;
+ box-sizing: border-box;
+ float: left;
+ margin-left: 2.762430939%;
+ *margin-left: 2.709239449638298%;
+ }
+ .row-fluid [class*="span"]:first-child {
+ margin-left: 0;
+ }
+ .row-fluid .span12 {
+ width: 99.999999993%;
+ *width: 99.9468085036383%;
+ }
+ .row-fluid .span11 {
+ width: 91.436464082%;
+ *width: 91.38327259263829%;
+ }
+ .row-fluid .span10 {
+ width: 82.87292817100001%;
+ *width: 82.8197366816383%;
+ }
+ .row-fluid .span9 {
+ width: 74.30939226%;
+ *width: 74.25620077063829%;
+ }
+ .row-fluid .span8 {
+ width: 65.74585634900001%;
+ *width: 65.6926648596383%;
+ }
+ .row-fluid .span7 {
+ width: 57.182320438000005%;
+ *width: 57.129128948638304%;
+ }
+ .row-fluid .span6 {
+ width: 48.618784527%;
+ *width: 48.5655930376383%;
+ }
+ .row-fluid .span5 {
+ width: 40.055248616%;
+ *width: 40.0020571266383%;
+ }
+ .row-fluid .span4 {
+ width: 31.491712705%;
+ *width: 31.4385212156383%;
+ }
+ .row-fluid .span3 {
+ width: 22.928176794%;
+ *width: 22.874985304638297%;
+ }
+ .row-fluid .span2 {
+ width: 14.364640883%;
+ *width: 14.311449393638298%;
+ }
+ .row-fluid .span1 {
+ width: 5.801104972%;
+ *width: 5.747913482638298%;
+ }
+ input,
+ textarea,
+ .uneditable-input {
+ margin-left: 0;
+ }
+ input.span12, textarea.span12, .uneditable-input.span12 {
+ width: 714px;
+ }
+ input.span11, textarea.span11, .uneditable-input.span11 {
+ width: 652px;
+ }
+ input.span10, textarea.span10, .uneditable-input.span10 {
+ width: 590px;
+ }
+ input.span9, textarea.span9, .uneditable-input.span9 {
+ width: 528px;
+ }
+ input.span8, textarea.span8, .uneditable-input.span8 {
+ width: 466px;
+ }
+ input.span7, textarea.span7, .uneditable-input.span7 {
+ width: 404px;
+ }
+ input.span6, textarea.span6, .uneditable-input.span6 {
+ width: 342px;
+ }
+ input.span5, textarea.span5, .uneditable-input.span5 {
+ width: 280px;
+ }
+ input.span4, textarea.span4, .uneditable-input.span4 {
+ width: 218px;
+ }
+ input.span3, textarea.span3, .uneditable-input.span3 {
+ width: 156px;
+ }
+ input.span2, textarea.span2, .uneditable-input.span2 {
+ width: 94px;
+ }
+ input.span1, textarea.span1, .uneditable-input.span1 {
+ width: 32px;
+ }
+}
+@media (min-width: 1200px) {
+ .row {
+ margin-left: -30px;
+ *zoom: 1;
+ }
+ .row:before,
+ .row:after {
+ display: table;
+ content: "";
+ }
+ .row:after {
+ clear: both;
+ }
+ [class*="span"] {
+ float: left;
+ margin-left: 30px;
+ }
+ .container,
+ .navbar-fixed-top .container,
+ .navbar-fixed-bottom .container {
+ width: 1170px;
+ }
+ .span12 {
+ width: 1170px;
+ }
+ .span11 {
+ width: 1070px;
+ }
+ .span10 {
+ width: 970px;
+ }
+ .span9 {
+ width: 870px;
+ }
+ .span8 {
+ width: 770px;
+ }
+ .span7 {
+ width: 670px;
+ }
+ .span6 {
+ width: 570px;
+ }
+ .span5 {
+ width: 470px;
+ }
+ .span4 {
+ width: 370px;
+ }
+ .span3 {
+ width: 270px;
+ }
+ .span2 {
+ width: 170px;
+ }
+ .span1 {
+ width: 70px;
+ }
+ .offset12 {
+ margin-left: 1230px;
+ }
+ .offset11 {
+ margin-left: 1130px;
+ }
+ .offset10 {
+ margin-left: 1030px;
+ }
+ .offset9 {
+ margin-left: 930px;
+ }
+ .offset8 {
+ margin-left: 830px;
+ }
+ .offset7 {
+ margin-left: 730px;
+ }
+ .offset6 {
+ margin-left: 630px;
+ }
+ .offset5 {
+ margin-left: 530px;
+ }
+ .offset4 {
+ margin-left: 430px;
+ }
+ .offset3 {
+ margin-left: 330px;
+ }
+ .offset2 {
+ margin-left: 230px;
+ }
+ .offset1 {
+ margin-left: 130px;
+ }
+ .row-fluid {
+ width: 100%;
+ *zoom: 1;
+ }
+ .row-fluid:before,
+ .row-fluid:after {
+ display: table;
+ content: "";
+ }
+ .row-fluid:after {
+ clear: both;
+ }
+ .row-fluid [class*="span"] {
+ display: block;
+ width: 100%;
+ min-height: 28px;
+ -webkit-box-sizing: border-box;
+ -moz-box-sizing: border-box;
+ -ms-box-sizing: border-box;
+ box-sizing: border-box;
+ float: left;
+ margin-left: 2.564102564%;
+ *margin-left: 2.510911074638298%;
+ }
+ .row-fluid [class*="span"]:first-child {
+ margin-left: 0;
+ }
+ .row-fluid .span12 {
+ width: 100%;
+ *width: 99.94680851063829%;
+ }
+ .row-fluid .span11 {
+ width: 91.45299145300001%;
+ *width: 91.3997999636383%;
+ }
+ .row-fluid .span10 {
+ width: 82.905982906%;
+ *width: 82.8527914166383%;
+ }
+ .row-fluid .span9 {
+ width: 74.358974359%;
+ *width: 74.30578286963829%;
+ }
+ .row-fluid .span8 {
+ width: 65.81196581200001%;
+ *width: 65.7587743226383%;
+ }
+ .row-fluid .span7 {
+ width: 57.264957265%;
+ *width: 57.2117657756383%;
+ }
+ .row-fluid .span6 {
+ width: 48.717948718%;
+ *width: 48.6647572286383%;
+ }
+ .row-fluid .span5 {
+ width: 40.170940171000005%;
+ *width: 40.117748681638304%;
+ }
+ .row-fluid .span4 {
+ width: 31.623931624%;
+ *width: 31.5707401346383%;
+ }
+ .row-fluid .span3 {
+ width: 23.076923077%;
+ *width: 23.0237315876383%;
+ }
+ .row-fluid .span2 {
+ width: 14.529914530000001%;
+ *width: 14.4767230406383%;
+ }
+ .row-fluid .span1 {
+ width: 5.982905983%;
+ *width: 5.929714493638298%;
+ }
+ input,
+ textarea,
+ .uneditable-input {
+ margin-left: 0;
+ }
+ input.span12, textarea.span12, .uneditable-input.span12 {
+ width: 1160px;
+ }
+ input.span11, textarea.span11, .uneditable-input.span11 {
+ width: 1060px;
+ }
+ input.span10, textarea.span10, .uneditable-input.span10 {
+ width: 960px;
+ }
+ input.span9, textarea.span9, .uneditable-input.span9 {
+ width: 860px;
+ }
+ input.span8, textarea.span8, .uneditable-input.span8 {
+ width: 760px;
+ }
+ input.span7, textarea.span7, .uneditable-input.span7 {
+ width: 660px;
+ }
+ input.span6, textarea.span6, .uneditable-input.span6 {
+ width: 560px;
+ }
+ input.span5, textarea.span5, .uneditable-input.span5 {
+ width: 460px;
+ }
+ input.span4, textarea.span4, .uneditable-input.span4 {
+ width: 360px;
+ }
+ input.span3, textarea.span3, .uneditable-input.span3 {
+ width: 260px;
+ }
+ input.span2, textarea.span2, .uneditable-input.span2 {
+ width: 160px;
+ }
+ input.span1, textarea.span1, .uneditable-input.span1 {
+ width: 60px;
+ }
+ .thumbnails {
+ margin-left: -30px;
+ }
+ .thumbnails > li {
+ margin-left: 30px;
+ }
+ .row-fluid .thumbnails {
+ margin-left: 0;
+ }
+}
+@media (max-width: 979px) {
+ body {
+ padding-top: 0;
+ }
+ .navbar-fixed-top,
+ .navbar-fixed-bottom {
+ position: static;
+ }
+ .navbar-fixed-top {
+ margin-bottom: 18px;
+ }
+ .navbar-fixed-bottom {
+ margin-top: 18px;
+ }
+ .navbar-fixed-top .navbar-inner,
+ .navbar-fixed-bottom .navbar-inner {
+ padding: 5px;
+ }
+ .navbar .container {
+ width: auto;
+ padding: 0;
+ }
+ .navbar .brand {
+ padding-left: 10px;
+ padding-right: 10px;
+ margin: 0 0 0 -5px;
+ }
+ .nav-collapse {
+ clear: both;
+ }
+ .nav-collapse .nav {
+ float: none;
+ margin: 0 0 9px;
+ }
+ .nav-collapse .nav > li {
+ float: none;
+ }
+ .nav-collapse .nav > li > a {
+ margin-bottom: 2px;
+ }
+ .nav-collapse .nav > .divider-vertical {
+ display: none;
+ }
+ .nav-collapse .nav .nav-header {
+ color: #999999;
+ text-shadow: none;
+ }
+ .nav-collapse .nav > li > a,
+ .nav-collapse .dropdown-menu a {
+ padding: 6px 15px;
+ font-weight: bold;
+ color: #999999;
+ -webkit-border-radius: 3px;
+ -moz-border-radius: 3px;
+ border-radius: 3px;
+ }
+ .nav-collapse .btn {
+ padding: 4px 10px 4px;
+ font-weight: normal;
+ -webkit-border-radius: 4px;
+ -moz-border-radius: 4px;
+ border-radius: 4px;
+ }
+ .nav-collapse .dropdown-menu li + li a {
+ margin-bottom: 2px;
+ }
+ .nav-collapse .nav > li > a:hover,
+ .nav-collapse .dropdown-menu a:hover {
+ background-color: #222222;
+ }
+ .nav-collapse.in .btn-group {
+ margin-top: 5px;
+ padding: 0;
+ }
+ .nav-collapse .dropdown-menu {
+ position: static;
+ top: auto;
+ left: auto;
+ float: none;
+ display: block;
+ max-width: none;
+ margin: 0 15px;
+ padding: 0;
+ background-color: transparent;
+ border: none;
+ -webkit-border-radius: 0;
+ -moz-border-radius: 0;
+ border-radius: 0;
+ -webkit-box-shadow: none;
+ -moz-box-shadow: none;
+ box-shadow: none;
+ }
+ .nav-collapse .dropdown-menu:before,
+ .nav-collapse .dropdown-menu:after {
+ display: none;
+ }
+ .nav-collapse .dropdown-menu .divider {
+ display: none;
+ }
+ .nav-collapse .navbar-form,
+ .nav-collapse .navbar-search {
+ float: none;
+ padding: 9px 15px;
+ margin: 9px 0;
+ border-top: 1px solid #222222;
+ border-bottom: 1px solid #222222;
+ -webkit-box-shadow: inset 0 1px 0 rgba(255,255,255,.1), 0 1px 0 rgba(255,255,255,.1);
+ -moz-box-shadow: inset 0 1px 0 rgba(255,255,255,.1), 0 1px 0 rgba(255,255,255,.1);
+ box-shadow: inset 0 1px 0 rgba(255,255,255,.1), 0 1px 0 rgba(255,255,255,.1);
+ }
+ .navbar .nav-collapse .nav.pull-right {
+ float: none;
+ margin-left: 0;
+ }
+ .nav-collapse,
+ .nav-collapse.collapse {
+ overflow: hidden;
+ height: 0;
+ }
+ .navbar .btn-navbar {
+ display: block;
+ }
+ .navbar-static .navbar-inner {
+ padding-left: 10px;
+ padding-right: 10px;
+ }
+}
+@media (min-width: 980px) {
+ .nav-collapse.collapse {
+ height: auto !important;
+ overflow: visible !important;
+ }
+}
diff --git a/tutorial/template.class.html b/tutorial/template.class.html
new file mode 100644
index 00000000..44698c5d
--- /dev/null
+++ b/tutorial/template.class.html
@@ -0,0 +1,65 @@
+<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
+<html xmlns="http://www.w3.org/1999/xhtml"$if(lang)$ lang="$lang$" xml:lang="$lang$"$endif$>
+<head>
+ <meta http-equiv="Content-Type" content="text/html; charset=utf-8" />
+ <meta http-equiv="Content-Style-Type" content="text/css" />
+ <meta name="generator" content="pandoc" />
+$for(author-meta)$
+ <meta name="author" content="$author-meta$" />
+$endfor$
+$if(date-meta)$
+ <meta name="date" content="$date-meta$" />
+$endif$
+ <title>$if(title-prefix)$$title-prefix$ - $endif$$pagetitle$</title>
+ <style type="text/css">code{white-space: pre;}</style>
+$if(quotes)$
+ <style type="text/css">q { quotes: "“" "â€" "‘" "’"; }</style>
+$endif$
+$if(highlighting-css)$
+ <style type="text/css">
+$highlighting-css$
+ </style>
+$endif$
+$for(css)$
+ <link rel="stylesheet" href="$css$" $if(html5)$$else$type="text/css" $endif$/>
+$endfor$
+$if(math)$
+ $math$
+$endif$
+$for(header-includes)$
+ $header-includes$
+$endfor$
+</head>
+<body>
+ $if(title)$
+ <div class="navbar navbar-static-top">
+ <div class="navbar-inner">
+ <div class="container">
+ <span class="doc-title">$title$</span>
+ <ul class="nav pull-right doc-info">
+ $for(author)$
+ <li><p class="navbar-text">$author$</p></li>
+ $endfor$
+ $if(date)$
+ <li><p class="navbar-text">$date$</p></li>
+ $endif$
+ </ul>
+ </div>
+ </div>
+ </div>
+ $endif$
+ <div class="container">
+ <div class="row">
+ <div class="span12">
+ $for(include-before)$
+ $include-before$
+ $endfor$
+ $body$
+ $for(include-after)$
+ $include-after$
+ $endfor$
+ </div>
+ </div>
+ </div>
+</body>
+</html>
--
GitLab